2016 General Notebook
Author: Andrew D. Nguyen
Affiliation: Biology Department, University of Vermont
Contact: anbe642@gmail.com
Date started: 2016-05-13
Date end (last modified): 2017-01-01
Introduction:
I wish I started an online notebook earlier, but maybe it's not too late? Anyway, I'll use this doc to share my ideas and log the progress of my dissertation.

This work is licensed under a Creative Commons Attribution 4.0 International License.
General Lab protocols found here for heat shocks and RNA related experiments and here for protein related experiments.
Table of Contents (Layout follows Page number: Date. Title of entry)
Page 1: 2016-05-13. Indirect genetic effects
Page 2: 2016-05-13. Comparing G matrices of different populations
Page 3: 2016-05-16. Complete ddRAD-seq samples: processing
Page 4: 2016-05-16. Aphaenogaster morphological IDs
Page 5: 2016-05-16. Sequencing qPCR amplicons; Curtis and ANBE experiments
Page 6: 2016-05-17. Phylogenetics results from 2016-05-16 (CIPRES RaxML analysis)
Page 7: 2016-05-17. ABI steponeplus machine maintenance.
Page 8: 2016-05-18. Phylogenetic results excluding pogo (CIPRES RAXML analysis)
Page 9: 2016-05-18. Agarose gel electrophoresis of qPCR amplicons; Curtis and ANBE samples
Page 10: 2016-05-18. RaxML ML pairwise distance matrix
Page 11: 2016-05-18. ABI steponeplus machine maintenance update
Page 12: 2016-05-19. Getting whole rad loci with pyRAD and/or population in Stacks
Page 13: 2016-05-20. Evolution of proteome stability project
Page 14: 2016-05-24. Evolution of proteome stability project: Polyacrylamide gels for colony level replicates (A. rudis vs P. barbatus)
Page 15: 2016-05-24. Degenerate Hsp primer design from 2015-05-28
Page 16: 2016-05-24. Sequencing analysis continued from Page 5: 2016-05-16.
Page 17: 2016-05-25. Double check samples for SHC; JSG phytotron exp and MS.
Page 18: 2016-05-31. Learning model selection and model averaging!
- 2016-06-01: SHC suggestion. Just adding all phylo components in a global model instead of screening them out initially
- 2016-06-01: SHC suggestion. Just adding all phylo components in a global model instead of screening them out initially
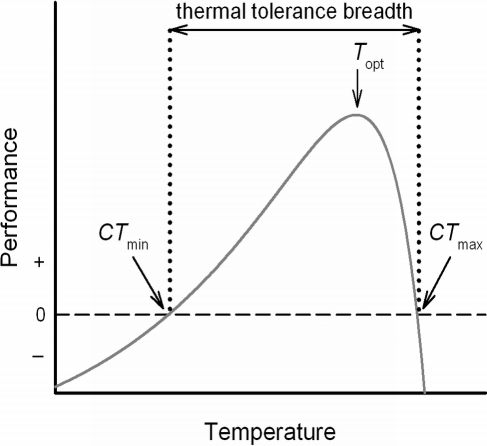
Page 19: 2016-06-01. Variance partitioning: thermal tolerance breadth example
Page 20: 2016-06-02. Notes from climate cascade meeting (2016-06-01)
Page 21: 2016-06-02. Levine's test for raw residuals
Page 22: 2016-06-02. Brute force fitting nls() functions in R!!
Page 23: 2016-06-02. Literature reference for thermal niche paper to help write manuscript
Page 24: 2016-06-03. Proteome stability project: Organizational entry
Page 25: 2016-06-03. ggplot reference, updating a figure from Page 20: 2016-06-02
Page 26: 2016-06-03. What is a cell type?
Page 27: 2016-06-03. qPCR plate layout and using the loaner ABI steponeplus Page 11: 2016-05-18
Page 28: 2016-06-03. Papers showing differences between fast static vs slow dynamic temperature treatments.
Page 29: 2016-06-06. Isolating RNA: colony CJ8; showing Sylvia
Page 30: 2016-06-07. Brute force fitting nls function in R revisited Page 22: 2016-06-02
[Failed attempt with nls2()] (#id-section30.1).
Page 31: 2016-06-08. Re-doing online notebook template
Page 32: 2016-06-08. qPCRs, 18s rRNA for Duke2, HF2, Kite 4, Kite8, 60 C annealing. Dilutions of future samples
Page 33: 2016-06-08. Climate cacade meeting
Page 34: 2016-06-09; 2016-06-10. qPCRs: Duke1, CJ2, SHC8, CJ5
Page 35: 2016-06-10. ABI steponeplus machine fix and sending back instrument.
Page 36: 2016-06-10. Thoughts on Kingsolver & Woods 2016, AmNat
- 2016-06-11. Follow up model
- 2016-06-13. Predictions of thermostat model
- 2016-06-11. Follow up model
Page 37: 2016-06-11. Quantifying natural selection in natural populations
Page 38: 2016-06-13. qPCR update for Duke1,CJ2,SHC8,CJ5. Randomizing samples treated at 25C(reference for basal expression) for qpcrs.
Page 39: 2016-06-13. Post doc project idea: Assessing current impacts of climate change in natural populations.
Page 40: 2016-06-14. qPCR's: Diluting samples for quantifying basal expression and repeats
Page 41: 2016-06-15. qPCRs to quantify basal expression (Evolution of stress response project)
Page 42: 2016-06-15. Evolution talks I want to attend.
Page 43: 2016-06-16. Figure for curve fitting: see Success with failwith() and Status update of samples.
Page 44: 2016-07-18. Summary statistics for modulation of Hsp paper.
Page 45: 2016-07-19. Meeting with VGN proteomics facility
Page 46: 2016-07-21. Reference samples for mapping index; Hsp modulation and thermal niche paper
Page 47: 2016-07-26. Learning mixed effects stat models
Page 48: 2016-07-27. Meeting with Steve Keller to discuss post doc idea (started here: Page 37: 2016-06-11. Quantifying natural selection in natural populations )
Page 49: 2016-07-28. Quantitative genetics and the molecular basis of complex traits
Page 50: 2016-08-02. Picking a plant system for post doc idea
Page 51: 2016-08-02; 2016-08-03. Climate cascade meeting
Page 52: 2016-08-04. Following up stats, range limits project
Page 53: 2016-08-08. Post doc ideas part 2 ; see Page 50: 2016-08-02. Picking a plant system for post doc idea
Page 54: 2016-08-10. Climate cascade meeting
Page 55: 2016-08-11. Overlaying raster files in a map in R
Page 56: 2016-08-16. range limits paper, data analysis of chill coma recovery time (CCRT) revisited
Page 57: 2016-08-25. Hsp modulation follow up stats
Page 58: 2016-08-29 and 30. Climate cascade meeting
Page 59: 2016-09-01. SHC lab meeting Fall 2016
Page 60: 2016-09-01. Paper notes: Paccard A, Van Buskirk J, Willi Y, Eckert CG, Bronstein JL. 2016. Quantitative Genetic Architecture at Latitudinal Range Boundaries: Reduced Variation but Higher Trait Independence. The American Naturalist.
Page 61: 2016-09-06. Playing with rpart with range limit data
Page 62: 2016-09-06. Climate cascade meeting
Page 63: 2016-09-07. PCA update for range limit data; see Page 61: 2016-09-06. Playing with rpart with range limit data
Page 64: 2016-09-12. ref for time table, nsf post doc grant
Page 65: 2016-09-12. variable importance
Page 66: 2016-09-13. climate cascade meeting
Page 67: 2016-09-14. SICB meeting
Page 68: 2016-09-19; 2016-09-20. Climate cascade meeting
Page 69: 2016-09-21. qpcr redos for 18s rRNA
Page 70: 2016-09-26. selecting poplar clones
Page 71: 2016-09-26 and 2016-09-27. Climate cascade meeting
Page 72: 2016-09-27 . evolution of hsp gxp data analysis
Page 73: 2016-09-28. building ultrametric trees
Page 74: 2016-09-28. phylogenetic regressions (PGLS) and anovas
- 2016-09-28. SHC suggestion: ancestral trait reconsturction -> regressions/anovas; following this tutorial
- 2016-09-29. PIC
Page 75: 2016-10-03 and 2016-10-04. Climate cascade meeting
Page 76: 2016-10-03 and 2016-10-04. Membrane stability
Page 77: 2016-10-04. Lab Safety Officer (LSO) meeting.
Page 78: 2016-10-05. Hsp gxp function valued trait fig
Page 79: 2016-10-06. SHC lab meeting: NSF post doc app
Page 80: 2016-10-07. Prepping cliamte cascade meeting
Page 81: 2016-10-11. ANCOVA models for testing interaction of hsp gxp parameter and habitat on CTmax
Page 82: 2016-10-11. variance partitioning in CTmax of aphaeno
Page 83: 2016-10-12. Testing effect of MAT on Hsp gxp and looking at correlations between phylogeny and climate.
Page 84: 2016-10-14. Updating climate cascade to do list.
Page 85: 2016-10-14. Paper note: Puentes, A., G. Granath, and J. Ågren. 2016. Similarity in G matrix structure among natural populations of Arabidopsis lyrata. Evolution 70:2370–2386.
Page 87: 2016-10-14. NSF post doc app meeting: Keller Lab
Page 88: 2016-10-18. Climate cascade meeting
Page 89: 2016-10-25. Climate cascade updated list
Page 90: 2016-10-25. Meeting with M Pespeni on 2016-10-26 and Brent 2016-10-27
Page 91: 2016-10-26. SICB meeting talk
Page 92: 2016-10-27. Proteome stability project update
Page 93: 2016-10-31. CTmax and Hsp reaction norm stats
Page 94: 2016-10-31; 2016-11-01. Climate cascade meeting setup and notes
Page 95: 2016-11-02. Ancestral trait reconstruction and CTmax PGLS ANBE common garden; corresponds with Page 74: 2016-09-28. phylogenetic regressions (PGLS) and anovas
Page 96: 2016-11-03. notes from skype meeting with KG, potential post doc opp
Page 97: 2016-11-04. ms in prep
Page 98: 2016-11-08. climate cascade meeting
Page 99: 2016-11-08. writing session with NJG and stats follow up
Page 100: 2016-11-14 & 2016-11-15. climate cascade meeting
Page 101: 2016-11-16. Hsp reaction norm stats; adding quadratic term
Page 102: 2016-11-22. climate cascade to do list
Page 103: 2016-12-06. climate cascade update
Page 104: 2016-12-19. climate cascade update
Page 105: 2016-12-20. Reading a few papers
Page 1: 2016-05-13. Indirect genetic effects.
Q:How does the social environment impact traits of individuals? Or what is the contribution of indirect genetic effects on an individual?
In ant colonies, sisters are highly related if the queen mated once.
H1: Ant workers traits are more optimal when the rearing environment is of the same genotype compared to different genotype.
Experiment: Cross foster experiment. Each ant colony is a different genotype, take 20 ants and split them up so each colony rears each other's babys.
This isnt a new idea: Linksvayer 2007. What would be interesting is to test the role of IGE in thermal ecology. Take a Northern(experiecnes cold) ant species and Southern (experiences warm) ant species and do a cross foster experiment. One outcome is that ants reared in the warm tolerant species will reare young in a way so that the baby has greater thermal tolerance than being reared by its own (cold tolerant genotype/species).
Page 2: 2016-05-13. Comparing G matrices of different populations
Since I've been an RA since January 2015, I've been able to teach myself things. One of my emerging obsessions is understanding how multiple traits evolve or respond to selection. For example a thermal performance curve is multivariate and how can this curve change?
It can vary vertically, shift right to left (warmer-cooler variation), and/or exhibit generalist-specialist variation. Kingsolver et al. 2105 has a cool paper showing how you can construct a G matrix, decompose it with a PCA to look at the genetic correlations and it subsequently captures how G matrices can change or thermal performance curves can respond to selection together. So all positive loadings equals vertical shifts, positive relationships of loadings with temperature equals warmer-cooler variation, and a bell shaped curve equals the generalist-specialist variation.
Example table of loadings across each temperature:
| Variation | 15 | 20 | 25 | 30 | 35 |
|---|---|---|---|---|---|
| Vertical | 1 | 1 | 1 | 1 | 1 |
| Warmer-cooler | -1 | -.5 | 0 | .5 | 1 |
| Generalist-specialist | -1 | .5 | 1 | .5 | -1 |
Whoa, what if you wanted to compare G matrices of different populations? One way is to do a PCA decomp with each G matrix constructed from each population. Then simply look at how the loadings change as a function of temperature between populations. Statistically, you can do an ANCOVA such as:
#Loadings is a continuous variable#Temperature can be a factor or continuous#Population is a factoraov(Loadings ~ Temperature * Population) A cool paper by Berger et al. 2013 has sort of done this(with out the ANCOVA). In table 3, they have gmax loadings(1st eigenvector of their G matrix) for each temperature for 3 populations: Norht, Central, South. So the Northern population exhibits warmer-cooler variation (high loadings low temps, negative loadings on high temps), whereas, Central and South exhibit vertical variation (all loadings are positive).
There is another cool paper to read about comparing G matrices by Aguirre et al. 2014.
- Random skewers method; simulate repsonse to selection by calculating it with randomized betas
- Common subspace; no clue what this is
- Construct a tensor; sounds like a 3D G matrix
- Decompose G into eigenvectors; like Kingsolver, I believe
Page 3: 2016-05-16. Complete ddRAD-seq samples: processing
ddrad-seq data are in! SHC processed short reads in STACKS and produced a fasta file.
From SHC:
*Hi Andrew, I have run all your samples against your index and through the STACKs pipeline - I used a minimum threshold of 5 reads to call a SNP, a maximum # of SNPs per tag of 6, and a minimum number of individuals that had to have a genotype call at a SNP of 10 individuals. The stats of genotype calls and heterozygosity across all your samples is in the excel spreadsheet - I highlighted those with <25% calls in yellow, and would not use those because they mess up the polarity inference for the SNPs and make the tree more ambiguous. The exception would probably be NOVCOC, since you will need an outgroup and none of the putative outgroup taxa meet the threshold. I've attached a NJ tree using all the >
25% taxa plus NOVCOC, and it seems to resolve very nicely bootstrap-wise. I do not know what many of these samples are, so no clue if it is biologically reasonable.
You'll find your fasta file in my scratch space here: Andrew_RADseq051516/final_Andrew_sam_files/m5output_refmap/Andrew_SNP_sequences_m5filter6ind10.fas Sara*
But, samples need to be redone:
*Hi Andrew, Just realized I did not adjust the barcode key for two samples in ddRAD10 that got moved during library prep – KITE5 and GF34-1. So their data are incorrect. Fixing now and should have a new version in a day or two. Sara*
So the following files should be disregarded but I'm keeping them just to log them:
In the 2014xanbe-common-garden_gxp_evolution/Data/Phylogenetics/20160516complete_dataset_phylo_analyses/
- 20160516SHC_Andrew_final_m5filter6ind10NJtree.pdf
- 20160516_SHC_Andrew_het-summary_SNPs.xlsx
- 20160516SHC_Andrew_SNP_sequences_m5filter6_ind10.fas
But if you ignore KITE5 and GF34-1, here is the summary of results:
| Sample | SNPs | Hets | Total | Prop.SNPs | Prop.het |
|---|---|---|---|---|---|
| FORMICA | 47 | 1 | 173822 | 0.00 | 0.021 |
| PB17-10_cat | 203 | 0 | 173822 | 0.00 | 0.000 |
| CAMPNSP | 584 | 25 | 173822 | 0.00 | 0.043 |
| PB17-14 | 1031 | 10 | 173822 | 0.01 | 0.010 |
| PB07-23 | 1921 | 14 | 173822 | 0.01 | 0.007 |
| 09A | 2587 | 32 | 173822 | 0.01 | 0.012 |
| CREMATOGASTER_cat | 3094 | 32 | 173822 | 0.02 | 0.010 |
| Kite8r | 3751 | 56 | 173822 | 0.02 | 0.015 |
| TU64_cat | 5217 | 45 | 173822 | 0.03 | 0.009 |
| Sal13-14r | 7905 | 78 | 173822 | 0.05 | 0.010 |
| BK6-1 | 10743 | 182 | 173822 | 0.06 | 0.017 |
| EXIT65 | 11612 | 120 | 173822 | 0.07 | 0.010 |
| NOVCOC1 | 12013 | 34 | 173822 | 0.07 | 0.003 |
| ALA4 | 18707 | 494 | 173822 | 0.11 | 0.026 |
| KITE4_cat | 36845 | 494 | 173822 | 0.21 | 0.013 |
| AHF3r | 39557 | 455 | 173822 | 0.23 | 0.012 |
| Duke3r | 53391 | 827 | 173822 | 0.31 | 0.015 |
| FBR5r | 61628 | 1072 | 173822 | 0.35 | 0.017 |
| KITE5_cat | 65777 | 1745 | 173822 | 0.38 | 0.027 |
| KH1 | 69951 | 977 | 173822 | 0.40 | 0.014 |
| KH2r | 72601 | 1021 | 173822 | 0.42 | 0.014 |
| BSK5r | 73690 | 1573 | 173822 | 0.42 | 0.021 |
| FBRAGG1 | 76194 | 830 | 173822 | 0.44 | 0.011 |
| AHW7 | 76776 | 1298 | 173822 | 0.44 | 0.017 |
| AHF1r | 77515 | 1038 | 173822 | 0.45 | 0.013 |
| KH3 | 78618 | 1099 | 173822 | 0.45 | 0.014 |
| Avon19-1 | 78942 | 1001 | 173822 | 0.45 | 0.013 |
| Avon19-3 | 80182 | 1137 | 173822 | 0.46 | 0.014 |
| MA | 80584 | 1546 | 173822 | 0.46 | 0.019 |
| AHW2 | 80841 | 1405 | 173822 | 0.47 | 0.017 |
| FBRAGG3 | 81143 | 1103 | 173822 | 0.47 | 0.014 |
| AHF2 | 82047 | 1399 | 173822 | 0.47 | 0.017 |
| CJ2r | 82383 | 1026 | 173822 | 0.47 | 0.012 |
| SHC2 | 84679 | 1541 | 173822 | 0.49 | 0.018 |
| CJ4 | 84824 | 1375 | 173822 | 0.49 | 0.016 |
| HW10 | 85989 | 1521 | 173822 | 0.49 | 0.018 |
| SHC9r | 87346 | 1526 | 173822 | 0.50 | 0.017 |
| MIC2 | 88435 | 1198 | 173822 | 0.51 | 0.014 |
| LPR4 | 90037 | 1529 | 173822 | 0.52 | 0.017 |
| DUKE2 | 91310 | 1890 | 173822 | 0.53 | 0.021 |
| Ala5r | 91524 | 2161 | 173822 | 0.53 | 0.024 |
| SHC10 | 91772 | 1614 | 173822 | 0.53 | 0.018 |
| CJ6r | 94419 | 1386 | 173822 | 0.54 | 0.015 |
| CJ7 | 95005 | 2888 | 173822 | 0.55 | 0.030 |
| LexSHC7r | 96193 | 1810 | 173822 | 0.55 | 0.019 |
| YATES1 | 96271 | 1921 | 173822 | 0.55 | 0.020 |
| DUKE1 | 96675 | 1731 | 173822 | 0.56 | 0.018 |
| SWSR45-1r | 97057 | 652 | 173822 | 0.56 | 0.007 |
| CJ8r | 99904 | 1318 | 173822 | 0.57 | 0.013 |
| LexSHC8r | 102414 | 1934 | 173822 | 0.59 | 0.019 |
| SHC5 | 102824 | 1916 | 173822 | 0.59 | 0.019 |
| SHC3 | 102969 | 1891 | 173822 | 0.59 | 0.018 |
| LEX9 | 103046 | 990 | 173822 | 0.59 | 0.010 |
| CJ3r | 103819 | 2001 | 173822 | 0.60 | 0.019 |
| ALA1_cat | 104644 | 2454 | 173822 | 0.60 | 0.023 |
| DUKE7 | 104763 | 3081 | 173822 | 0.60 | 0.029 |
| DUKE5 | 105184 | 2362 | 173822 | 0.61 | 0.022 |
| LPR1 | 105777 | 1459 | 173822 | 0.61 | 0.014 |
| LEX11 | 106302 | 1999 | 173822 | 0.61 | 0.019 |
| DUKE6 | 106634 | 1284 | 173822 | 0.61 | 0.012 |
| KH5 | 111245 | 1899 | 173822 | 0.64 | 0.017 |
| Avon19-2 | 111264 | 1667 | 173822 | 0.64 | 0.015 |
| Lex1r | 112200 | 2215 | 173822 | 0.65 | 0.020 |
| AHW4 | 112462 | 2571 | 173822 | 0.65 | 0.023 |
| KH7 | 113614 | 1765 | 173822 | 0.65 | 0.016 |
| NewSh20-2 | 114686 | 1843 | 173822 | 0.66 | 0.016 |
| KH6 | 116788 | 1914 | 173822 | 0.67 | 0.016 |
| Duke9r | 117894 | 1385 | 173822 | 0.68 | 0.012 |
| KH4 | 118160 | 1794 | 173822 | 0.68 | 0.015 |
| ALA3_cat | 118525 | 2965 | 173822 | 0.68 | 0.025 |
| CJ1 | 119737 | 1712 | 173822 | 0.69 | 0.014 |
| FBR4r | 122054 | 1894 | 173822 | 0.70 | 0.016 |
| Yates2r | 122085 | 2440 | 173822 | 0.70 | 0.020 |
| AHW1 | 122370 | 1423 | 173822 | 0.70 | 0.012 |
| YATES3 | 124183 | 2700 | 173822 | 0.71 | 0.022 |
| SHC6 | 124396 | 2577 | 173822 | 0.72 | 0.021 |
| Mon22-2 | 124452 | 2148 | 173822 | 0.72 | 0.017 |
| NP20-3 | 124543 | 2092 | 173822 | 0.72 | 0.017 |
| CJ9 | 124795 | 2533 | 173822 | 0.72 | 0.020 |
| Burn21-1 | 124846 | 2087 | 173822 | 0.72 | 0.017 |
| KH8 | 125663 | 2139 | 173822 | 0.72 | 0.017 |
| Can21-2 | 125727 | 2192 | 173822 | 0.72 | 0.017 |
| KITE1 | 126422 | 3578 | 173822 | 0.73 | 0.028 |
| GB33-1 | 127665 | 2376 | 173822 | 0.73 | 0.019 |
| CJ5r | 127719 | 2798 | 173822 | 0.73 | 0.022 |
| Duke8r | 128227 | 1555 | 173822 | 0.74 | 0.012 |
| SHC4r | 128586 | 2703 | 173822 | 0.74 | 0.021 |
| Ted3r | 129299 | 2332 | 173822 | 0.74 | 0.018 |
| TED4_cat | 131556 | 2828 | 173822 | 0.76 | 0.021 |
| Unit22-1 | 134451 | 2447 | 173822 | 0.77 | 0.018 |
| ALA2_cat | 134714 | 3708 | 173822 | 0.78 | 0.028 |
| Sap | 135261 | 2478 | 173822 | 0.78 | 0.018 |
| Pal21-3 | 135373 | 2400 | 173822 | 0.78 | 0.018 |
| POP2 | 135796 | 3030 | 173822 | 0.78 | 0.022 |
| Norr20-1 | 135922 | 2502 | 173822 | 0.78 | 0.018 |
| FBRAGG2 | 136534 | 3678 | 173822 | 0.79 | 0.027 |
| Duke4r | 136812 | 3048 | 173822 | 0.79 | 0.022 |
| Camb31-1 | 136979 | 2424 | 173822 | 0.79 | 0.018 |
| KITE2 | 137173 | 2190 | 173822 | 0.79 | 0.016 |
| Hamp23-1 | 137953 | 2639 | 173822 | 0.79 | 0.019 |
| LEX5 | 139853 | 3058 | 173822 | 0.80 | 0.022 |
| Pop1r | 139912 | 3187 | 173822 | 0.80 | 0.023 |
| GF34-1 | 140928 | 4088 | 173822 | 0.81 | 0.029 |
| POP3 | 140937 | 3175 | 173822 | 0.81 | 0.023 |
| LPR2 | 143401 | 2190 | 173822 | 0.82 | 0.015 |
| SHC1 | 145375 | 2371 | 173822 | 0.84 | 0.016 |
| AHW5 | 145662 | 2407 | 173822 | 0.84 | 0.017 |
| Phil20-4 | 147770 | 2915 | 173822 | 0.85 | 0.020 |
| AHW3 | 148236 | 3804 | 173822 | 0.85 | 0.026 |
| MIC1 | 149191 | 2737 | 173822 | 0.86 | 0.018 |
| LEX13 | 149260 | 3486 | 173822 | 0.86 | 0.023 |
| TED6 | 154029 | 3347 | 173822 | 0.89 | 0.022 |
| PMBE_cat | 163739 | 3120 | 173822 | 0.94 | 0.019 |
| KITE3 | 166928 | 6437 | 173822 | 0.96 | 0.039 |
Preliminary Tree; NJ:
SHC sent updated fasta file:
*Your fasta file should be ready again – turns out that GF34-1 mapped very poorly and really should not be used. The new SNP yield/heterozygosity summary file is in the same directory for you.
Sara*
Got rid of old fasta file, here is the updated file list:
- 20160516-Andrew_SNP_sequences_m5filter6ind10_het.tsv ; summary
- 20160516SHC_Andrew_SNP_sequences_m5filter6_ind10.fas ; unmodified names
- 20160516Andrew_SNP_sequences.fas; relabeled to match my sampling sheet; got rid of "trimmed90_filtered"
Summary table of updated fasta file:
| Sample | SNPs | Hets | Total | Proportion_loci_with_genotype |
|---|---|---|---|---|
| FORMICA | 43 | 1 | 174008 | 0.02 |
| PB17-10_cat | 203 | 0 | 174008 | 0.00 |
| CAMPNSP | 590 | 23 | 174008 | 0.04 |
| PB17-14 | 1034 | 9 | 174008 | 0.01 |
| PB07-23 | 1924 | 14 | 174008 | 0.01 |
| 09A | 2608 | 34 | 174008 | 0.01 |
| CREMATOGASTER_cat | 3087 | 32 | 174008 | 0.01 |
| Kite8r | 3688 | 53 | 174008 | 0.01 |
| GF34-1 | 4035 | 28 | 174008 | 0.01 |
| TU64_cat | 5180 | 45 | 174008 | 0.01 |
| Sal13-14r | 7892 | 78 | 174008 | 0.01 |
| BK6-1 | 10723 | 174 | 174008 | 0.02 |
| EXIT65 | 11573 | 122 | 174008 | 0.01 |
| NOVCOC1 | 12003 | 34 | 174008 | 0.00 |
| ALA4 | 18742 | 500 | 174008 | 0.03 |
| KITE4_cat | 36913 | 498 | 174008 | 0.01 |
| AHF3r | 39632 | 458 | 174008 | 0.01 |
| Duke3r | 53458 | 813 | 174008 | 0.02 |
| FBR5r | 61790 | 1073 | 174008 | 0.02 |
| KH1 | 70047 | 977 | 174008 | 0.01 |
| KH2r | 72760 | 1024 | 174008 | 0.01 |
| BSK5r | 73728 | 1575 | 174008 | 0.02 |
| FBRAGG1 | 76234 | 832 | 174008 | 0.01 |
| AHW7 | 76850 | 1278 | 174008 | 0.02 |
| AHF1r | 77526 | 1043 | 174008 | 0.01 |
| KH3 | 78767 | 1102 | 174008 | 0.01 |
| Avon19-1 | 79026 | 995 | 174008 | 0.01 |
| Avon19-3 | 80160 | 1125 | 174008 | 0.01 |
| MA | 80715 | 1536 | 174008 | 0.02 |
| AHW2 | 80937 | 1402 | 174008 | 0.02 |
| FBRAGG3 | 81176 | 1122 | 174008 | 0.01 |
| AHF2 | 82223 | 1396 | 174008 | 0.02 |
| CJ2r | 82528 | 1023 | 174008 | 0.01 |
| SHC2 | 84811 | 1527 | 174008 | 0.02 |
| CJ4 | 85003 | 1371 | 174008 | 0.02 |
| HW10 | 85935 | 1512 | 174008 | 0.02 |
| SHC9r | 87518 | 1514 | 174008 | 0.02 |
| MIC2 | 88542 | 1199 | 174008 | 0.01 |
| LPR4 | 90158 | 1530 | 174008 | 0.02 |
| DUKE2 | 91423 | 1896 | 174008 | 0.02 |
| Ala5r | 91632 | 2171 | 174008 | 0.02 |
| SHC10 | 91826 | 1595 | 174008 | 0.02 |
| CJ6r | 94504 | 1388 | 174008 | 0.01 |
| CJ7 | 95178 | 2898 | 174008 | 0.03 |
| LexSHC7r | 96265 | 1803 | 174008 | 0.02 |
| YATES1 | 96479 | 1934 | 174008 | 0.02 |
| DUKE1 | 96531 | 1570 | 174008 | 0.02 |
| SWSR45-1r | 97061 | 654 | 174008 | 0.01 |
| CJ8r | 100052 | 1315 | 174008 | 0.01 |
| LexSHC8r | 102556 | 1914 | 174008 | 0.02 |
| SHC5 | 102976 | 1895 | 174008 | 0.02 |
| LEX9 | 103074 | 994 | 174008 | 0.01 |
| SHC3 | 103077 | 1882 | 174008 | 0.02 |
| CJ3r | 103816 | 1963 | 174008 | 0.02 |
| ALA1_cat | 104771 | 2433 | 174008 | 0.02 |
| DUKE7 | 104940 | 3087 | 174008 | 0.03 |
| DUKE5 | 105313 | 2376 | 174008 | 0.02 |
| LPR1 | 105841 | 1459 | 174008 | 0.01 |
| LEX11 | 106390 | 1984 | 174008 | 0.02 |
| DUKE6 | 106792 | 1291 | 174008 | 0.01 |
| Avon19-2 | 111266 | 1661 | 174008 | 0.01 |
| KH5 | 111410 | 1902 | 174008 | 0.02 |
| Lex1r | 112257 | 2203 | 174008 | 0.02 |
| AHW4 | 112475 | 2552 | 174008 | 0.02 |
| KH7 | 113763 | 1762 | 174008 | 0.02 |
| NewSh20-2 | 114753 | 1863 | 174008 | 0.02 |
| KH6 | 116912 | 1917 | 174008 | 0.02 |
| Duke9r | 117978 | 1390 | 174008 | 0.01 |
| KH4 | 118263 | 1797 | 174008 | 0.02 |
| ALA3_cat | 118653 | 3003 | 174008 | 0.03 |
| CJ1 | 119837 | 1716 | 174008 | 0.01 |
| FBR4r | 122154 | 1887 | 174008 | 0.02 |
| Yates2r | 122241 | 2424 | 174008 | 0.02 |
| AHW1 | 122370 | 1435 | 174008 | 0.01 |
| YATES3 | 124252 | 2669 | 174008 | 0.02 |
| SHC6 | 124556 | 2553 | 174008 | 0.02 |
| Mon22-2 | 124561 | 2157 | 174008 | 0.02 |
| NP20-3 | 124747 | 2105 | 174008 | 0.02 |
| CJ9 | 124875 | 2508 | 174008 | 0.02 |
| Burn21-1 | 124936 | 2101 | 174008 | 0.02 |
| Can21-2 | 125784 | 2198 | 174008 | 0.02 |
| KH8 | 125792 | 2150 | 174008 | 0.02 |
| KITE1 | 126638 | 3576 | 174008 | 0.03 |
| GB33-1 | 127656 | 2385 | 174008 | 0.02 |
| CJ5r | 127851 | 2772 | 174008 | 0.02 |
| Duke8r | 128355 | 1556 | 174008 | 0.01 |
| SHC4r | 128604 | 2669 | 174008 | 0.02 |
| Ted3r | 129289 | 2348 | 174008 | 0.02 |
| TED4_cat | 131758 | 2863 | 174008 | 0.02 |
| Unit22-1 | 134508 | 2472 | 174008 | 0.02 |
| ALA2_cat | 134818 | 3729 | 174008 | 0.03 |
| Pal21-3 | 135398 | 2411 | 174008 | 0.02 |
| Sap | 135413 | 2487 | 174008 | 0.02 |
| POP2 | 135928 | 3004 | 174008 | 0.02 |
| Norr20-1 | 136013 | 2506 | 174008 | 0.02 |
| FBRAGG2 | 136626 | 3680 | 174008 | 0.03 |
| Duke4r | 136895 | 3035 | 174008 | 0.02 |
| Camb31-1 | 137074 | 2448 | 174008 | 0.02 |
| KITE2 | 137322 | 2185 | 174008 | 0.02 |
| Hamp23-1 | 138088 | 2646 | 174008 | 0.02 |
| Pop1r | 139982 | 3140 | 174008 | 0.02 |
| LEX5 | 139987 | 3014 | 174008 | 0.02 |
| POP3 | 141037 | 3140 | 174008 | 0.02 |
| LPR2 | 143432 | 2185 | 174008 | 0.02 |
| SHC1 | 145541 | 2382 | 174008 | 0.02 |
| AHW5 | 145766 | 2409 | 174008 | 0.02 |
| Phil20-4 | 147887 | 2925 | 174008 | 0.02 |
| AHW3 | 148314 | 3796 | 174008 | 0.03 |
| MIC1 | 149322 | 2762 | 174008 | 0.02 |
| LEX13 | 149401 | 3461 | 174008 | 0.02 |
| TED6 | 154109 | 3362 | 174008 | 0.02 |
| KITE5_cat | 157748 | 5246 | 174008 | 0.03 |
| PMBE_cat | 163881 | 3111 | 174008 | 0.02 |
| KITE3 | 167083 | 6441 | 174008 | 0.04 |
Parsed 20160516_Andrew_SNP_sequences.fas:
got rid of samples with low number of SNPs
- FORMICA
- PB17-10
- CAMPNSP
- PB17-14
- PB07-23
- 09A
- CREMATOGASTER
- Kite8r
- GF34-1
Grabbing number of samples from command line:
grep '^>' 20160516_Andrew_SNP_sequences.fas | wc -l
107
107 samples!
Next step is to reconstruct relationships of SNP Matrix
- Use CIPRES
- Use RAxML-HPC BlackBox (8.2.8) to reconstruct ML tree
- I also need to estimate the ML distance matrix with computer in ant room.
For ML distance matrix with raxml, you need a fasta file and tree. Piece of code I've tried before:
*##for anbe tree, claculate pairwise ml distance matrix nohup nice -n 19 ./raxmlHPC -f x -p 12345 -s ~/Desktop/2015ANBE_common_garden/20150818Andrew_SNP_sequences_nooutgr.fasta -m GTRGAMMA -t ~/Desktop/2015ANBE_common_garden/RAxML_bestTree.20150819commongarden_raxml_v2 -n 20150828_commongarden_pairwise_ML_distance &
Page 4: 2016-05-13. Aphaenogaster morphological IDs
For JSG phytotron project (and also partly Lchick's thermal niche paper).
| ID | Colony.ID | Species | Vouchers | Bernice.morphological.ID | pinned | sample.from | notes |
|---|---|---|---|---|---|---|---|
| ApGXL-01-A | MagSpr3 | carolinensis | no specimen | ||||
| ApGXL-01-B | MagSpr4 | rudis | no specimen | ||||
| ApGXL-01-C | MagSpr7 | carolinensis | no specimen | ||||
| ApGXL-02-A | HW1 | rudis | rudis | y | Clint | ||
| ApGXL-02-B | HW5 | rudis | no specimen | ||||
| ApGXL-02-C | HW7 | rudis | voucherNCSU | rudis | y | Clint | |
| ApGXL-03-A | FMU4 | . | no specimen | ||||
| ApGXL-04-A | UNF8 | rudis | rudis | n | Sara | ||
| ApGXL-04-B | UNF9 | rudis | rudis | n | Sara | ||
| ApGXL-04-C | UNF1 | carolinensis | carolinensis | n | Sara | ||
| ApGXL-05-B | GSMNP4 | picea | picea | y | Sara | ||
| ApGXL-05-D | GSMNP5 | picea | picea | y | Sara | ||
| ApGXL-06-A | DW2 | rudis | rudis | n | Clint | ||
| ApGXL-06-B | DW1 | rudis | rudis | n | Sara | ||
| ApGXL-07-A | BRP2 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-07-B | BRP9 | picea | voucherNCSU | no specimen | |||
| ApGXL-08-A | Ijams6 | rudis | rudis | y | Sara | ||
| ApGXL-08-D | IJams1 | rudis | rudis | n | Sara | ||
| ApGXL-09-A | RC12 | rudis | rudis | n | Clint | ||
| ApGXL-10-A | LVA9 | rudis | rudis | n | Sara | there are 2 LVA 9s, not sure which one I have | |
| ApGXL-10-B | LVA12 | rudis | rudis | n | Sara | ||
| ApGXL-10-C | LVA11 | fulva | fulva | n | Sara | ||
| ApGXL-10-F | LVA9 | rudis | rudis | n | Sara | there are 2 LVA 9s, not sure which one I have | |
| ApGXL-11-A | WP9 | rudis | voucherNCSU | rudis | y | Clint | |
| ApGXL-11-B | WP11 | rudis | voucherNCSU | rudis? | y | Clint | where is this from? It looks dark like picea, but no lighter antennal segmants. It resembles one I described morphologically as rudis, but DNA said picea |
| ApGXL-11-C | WP3 | fulva | voucherNCSU | fulva | y | Clint | |
| ApGXL-11-D | WP6 | rudis | rudis | n | Sara | ||
| ApGXL-12-A | NOCK6 | picea | rudis | n | Clint | where is this from? It looks dark like picea, but no lighter antennal segmants. It resembles one I described morphologically as rudis, but DNA said picea | |
| ApGXL-12-D | NOCK8 | rudis | rudis | y | Sara | where is this from? It looks dark like picea, but no lighter antennal segmants. It resembles one I described morphologically as rudis, but DNA said picea | |
| ApGXL-13-A | HSP6 | picea | picea | n | Sara | ||
| ApGXL-13-B | HSP7 | picea | picea | n | Sara | ||
| ApGXL-13-C | HSP9 | picea | voucherNCSU | picea | y | Clint | where is this from? It looks dark like picea, but no lighter antennal segmants. It resembles one I described morphologically as rudis, but DNA said picea |
| ApGXL-13-D | HSP12 | picea | picea | y | Sara | ||
| ApGXL-15-A | DSF4 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-15-B | DSF11 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-15-C | DSF8 | picea | picea | n | Sara | ||
| ApGXL-15-D | DSF12 | picea | voucherNCSU | picea | y | Clint | |
| APGXL-16-A | BRM4 | picea | picea | n | Sara | ||
| APGXL-16-B | BRM/BRF8 | picea | picea | n | Sara | ||
| ApGXL-17-A | Bard10 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-17-B | Bard9 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-17-C | Bard3 | picea | picea | n | Sara | ||
| ApGXL-18-A | Notch1 | fulva | voucherNCSU | picea | y | Sara | discrepancy - spines not upward |
| ApGXL-18-C | Notch4 | rudis | picea | n | Sara | discrepancy, last 4 antennal sements lighter in color) | |
| ApGXL-18-D | Notch2 | fulva | voucherNCSU | picea | y | Clint | discrepancy - spines not upward |
| ApGXL-19-A | HF001 | picea | picea | n | Sara | ||
| ApGXL-20-A | APB10 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-20-B | APB3a | picea | picea | n | Sara | ||
| ApGXL-20-C | APB3b | picea | picea | n | Sara | ||
| ApGXL-20-D | APB8 | picea | picea | n | Sara | ||
| ApGXL-21-A | Bear6 | picea | picea | n | Sara | ||
| ApGXL-21-B | Bear5 | picea | picea | n | Sara | ||
| ApGXL-21-C | Bear3 | picea | picea | y | Sara | ||
| ApGXL-22-A | SEB1 | . | picea | n | Sara | ||
| ApGXL-22-B | SEB8 | picea | picea | n | Sara | ||
| ApGXL-22-C | SEB9 | picea | picea | n | Sara | ||
| ApGXL-23-A | MM1 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-23-B | MM2 | picea | picea | n | Sara | ||
| ApGXL-23-C | MM4 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-24-A | EW09 | picea | picea | n | Sara | ||
| ApGXL-24-B | EW4 | . | picea | n | Sara | ||
| ApGXL-25-A | RW3 | picea | voucherNCSU | picea | y | Clint | light, but last 4 antennal segments lighter |
| ApGXL-25-C | RW1 | . | no specimen | ||||
| ApGXL-25-D | RW5 | picea | picea | n | Sara | ||
| ApGXL-26-A | MB1 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-26-B | MB3 | picea | voucherNCSU | no specimen | |||
| ApGXL-26-C | MB4 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-26-D | MB2 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-26-E | MB6 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-27-A | KBH4b | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-27-B | KBH1 | picea | voucherNCSU | picea | y | Clint | |
| ApGXL-28-A | Brad1 | picea | picea | y | Sara | ||
| ApGXL-28-B | Brad6 | picea | voucherNCSU | picea | y | Clint | |
| Aphaen 15 | Aphaen 15 | ||||||
| Aphaen A2 | Aphaen A2 | ||||||
| Aphaen12 | Aphaen12 | ||||||
| Aphaen17 | Aphaen17 | ||||||
| Aphaen18 | Aphaen18 | rudis | |||||
| AphaenA | AphaenA | rudis | voucherNCSU | ||||
| AphaenB | AphaenB | ||||||
| BARD11 | BARD11 | ||||||
| BARD2 | BARD2 | picea | |||||
| BARD5 | BARD5 | fulva | |||||
| Blank | Blank | rudis | voucherNCSU | ||||
| Brad2 | Brad2 | picea | voucherNCSU | ||||
| Brad3 | Brad3 | ||||||
| BRP-2B | BRP-2B | picea | |||||
| BRP08 | BRP08 | ||||||
| BRP1 | BRP1 | picea | voucherNCSU | ||||
| BRP10 | BRP10 | ||||||
| BRP11 | BRP11 | picea | voucherNCSU | ||||
| BRP3 | BRP3 | picea | voucherNCSU | ||||
| BRP5 | BRP5 | picea | voucherNCSU | ||||
| BRP6 | BRP6 | ||||||
| BRP7 | BRP7 | picea | voucherNCSU | ||||
| DF-3A | DF-3A | rudis | voucherNCSU | ||||
| DF1-A | DF1-A | rudis | voucherNCSU | ||||
| FMU6 | FMU6 | rudis | voucherNCSU | ||||
| HSP1 | HSP1 | picea | |||||
| HSP4 | HSP4 | ||||||
| HSP5 | HSP5 | picea | |||||
| HW8 | HW8 | ||||||
| HW9 | HW9 | ||||||
| KBH6 | KBH6 | ||||||
| KBH8 | KBH8 | picea | voucherNCSU | ||||
| LVA1 | LVA1 | fulva | voucherNCSU | ||||
| LVA13 | LVA13 | rudis | voucherNCSU | ||||
| LVA2 | LVA2 | rudis | voucherNCSU | ||||
| LVA3 | LVA3 | rudis | voucherNCSU | ||||
| MAGSPR6 | MAGSPR6 | rudis | voucherNCSU | ||||
| NSP2 | NSP2 | picea | voucherNCSU | ||||
| NSP3 | NSP3 | rudis | |||||
| NSP7 | NSP7 | fulva | |||||
| OLDRC1 | OLDRC1 | fulva | |||||
| OldRC3 | OldRC3 | fulva | |||||
| OldRC4 | OldRC4 | rudis | |||||
| OldRC6 | OldRC6 | rudis | |||||
| OLDRC7 | OLDRC7 | rudis | |||||
| RC02 | RC02 | fulva | voucherNCSU | ||||
| RC04 | RC04 | rudis | |||||
| RC06 | RC06 | rudis | voucherNCSU | ||||
| RC09 | RC09 | rudis | voucherNCSU | ||||
| RC10 | RC10 | rudis | voucherNCSU | ||||
| RC11 | RC11 | rudis | |||||
| RC13 | RC13 | rudis | voucherNCSU | ||||
| RC14 | RC14 | rudis | |||||
| RC15 | RC15 | rudis | |||||
| RC16 | RC16 | rudis | |||||
| Seb 2A | Seb 2A | ||||||
| SEB3A | SEB3A | ||||||
| UNF4A | UNF4A | rudis | |||||
| UNF7A | UNF7A | miamiana | |||||
| YM01 | YM01 | rudis | |||||
| YM02 | YM02 | rudis | |||||
| YM03 | YM03 | rudis |
Page 5: 2016-05-13. Sequencing qPCR amplicons; Curtis and ANBE experiments
Sample list and plate layout for sanger sequencing. Amplicons ~ 100bps and were Qiagen PCR purified following manufacturer's instructions. Added ~3 ng template,with 2 uM primer in 11.6 uL volume. Curtis' chamber samples are on here and my own ANBE gene expression experiment. Submitting to vermont cancer center.
If interested in protocols , see here.
| Well | Template.Name | Primer.Name |
|---|---|---|
| A1 | HF 5-1 | 18s_F328 |
| B1 | HF 5-1 | 18s_R427 |
| C1 | HF 7-1 | 18s_F328 |
| D1 | HF 7-1 | 18s_R427 |
| E1 | DF 13-A | 18s_F328 |
| F1 | DF 13-A | 18s_R427 |
| G1 | DF 14-A | 18s_F328 |
| H1 | DF 14-A | 18s_R427 |
| A2 | DF 8-B | hsp83_F1583 |
| B2 | DF 8-B | hsp83_R1682 |
| C2 | DF 5C-4 | hsp83_F1583 |
| D2 | DF 5C-4 | hsp83_R1682 |
| E2 | HF 8-1 | hsp83_F1583 |
| F2 | HF 8-1 | hsp83_R1682 |
| G2 | HF 2-2 | hsp83_F1583 |
| H2 | HF 2-2 | hsp83_R1682 |
| A3 | DF 1-D | hsp70_F1468 |
| B3 | DF 1-D | hsp70_R1592 |
| C3 | DF 10-3 | hsp70_F1468 |
| D3 | DF 10-3 | hsp70_R1592 |
| E3 | HF2 8-2 | hsp70_F1468 |
| F3 | HF2 8-2 | hsp70_R1592 |
| G3 | HF2 4-1 | hsp70_F1468 |
| H3 | HF2 4-1 | hsp70_R1592 |
| A4 | HF2 7-2 | hsp40_F541 |
| B4 | HF2 7-2 | hsp40_R641 |
| C4 | HF2 5-2 | hsp40_F541 |
| D4 | HF2 5-2 | hsp40_R641 |
| E4 | DF A1-B | hsp40_F541 |
| F4 | DF A1-B | hsp40_R641 |
| G4 | DF A8-B | hsp40_F541 |
| H4 | DF A8-B | hsp40_R641 |
| A5 | HF2 5-3 | actin_F984 |
| B5 | HF2 5-3 | actin_R1095 |
| C5 | HF2 8-1 | actin_F984 |
| D5 | HF2 8-1 | actin_R1095 |
| E5 | DF 3-A | actin_F984 |
| F5 | DF 3-A | actin_R1095 |
| G5 | DF 7-A | actin_F984 |
| H5 | DF 7-A | actin_R1095 |
| A6 | Exit65 | 70_1468 |
| B6 | BK | 70_1468 |
| C6 | Ted6 | 70_1468 |
| D6 | DUKE6 | 70_1468 |
| E6 | ALA1 | 70_1468 |
| F6 | KH2 | 70_1468 |
| G6 | FB2 | 70_1468 |
| H6 | Exit65 | 70_1592 |
| A7 | BK | 70_1592 |
| B7 | Ted6 | 70_1592 |
| C7 | DUKE6 | 70_1592 |
| D7 | ALA1 | 70_1592 |
| E7 | KH2 | 70_1592 |
| F7 | FB2 | 70_1592 |
| G7 | Exit65 | 83_1583 |
| H7 | BK | 83_1583 |
| A8 | TED3 | 83_1583 |
| B8 | DUKE6 | 83_1583 |
| C8 | ALA1 | 83_1583 |
| D8 | KH2 | 83_1583 |
| E8 | FB2 | 83_1583 |
| F8 | Exit65 | 83_1682 |
| G8 | BK | 83_1682 |
| H8 | TED3 | 83_1682 |
| A9 | DUKE6 | 83_1682 |
| B9 | ALA1 | 83_1682 |
| C9 | KH2 | 83_1682 |
| D9 | FB2 | 83_1682 |
| E9 | PB1710 | 83_279 |
| F9 | POP2 | 83_279 |
| G9 | SHC2 | 83_279 |
| H9 | cremato | 83_279 |
| A10 | ex | 83_279 |
| B10 | bk | 83_279 |
| C10 | TED6 | 83_279 |
| D10 | PB1710 | 83_300 |
| E10 | POP2 | 83_300 |
| F10 | SHC2 | 83_300 |
| G10 | cremato | 83_300 |
| H10 | ex | 83_300 |
| A11 | bk | 83_300 |
| B11 | TED6 | 83_300 |
| C11 | DUKE6 | hsp40_541 |
| D11 | ALA1 | hsp40_541 |
| E11 | KH2 | hsp40_541 |
| F11 | FB2 | hsp40_541 |
| G11 | EX | hsp40_541 |
| H11 | BK | hsp40_541 |
| A12 | Ted6 | hsp40_541 |
| B12 | DUKE6 | hsp40_641 |
| C12 | ALA1 | hsp40_641 |
| D12 | KH2 | hsp40_641 |
| E12 | FB2 | hsp40_641 |
| F12 | EX | hsp40_641 |
| G12 | BK | hsp40_641 |
| H12 | Ted6 | hsp40_641 |
Page 6: 2016-05-17 Phylogenetics results from 2016-05-16 (CIPRES RaxML analysis)
Results from 2016-05-16 ML tree using RaxML black box on CIPRES.
Transformed branch lengths

Untransformed branch lengths

Notes: I left a pogo sample in there. LPR4 and HW5 look switched.
Summary of tree by species:

When comparing with the NJ tree, the placement of A. picea is different.
- ML tree: A. picea is sister to A. rudis,A. miamiana, A. lamellidens
- NJ tree: A. picea is sister to A. rudis,A. miamiana, A. lamellidens,A. ashmeadi, A. floridana
Rerunning analysis without PB07-23 to double check this sample doesnt skew ingroup relationships.
Page 7: 2016-05-17. ABI steponeplus machine maintenance.
Machine Problem: It freezes mid run without giving an error, even while operating stand alone. Sometimes when it freezes, the door wont release plate. And it also has trouble connecting to laptop even after restart.
Machine Info: ABI steponeplus
- serial #: 272007769
- ref: 4376592
- University #: A92219
Under contract, no cost.
Contact info:
- Jeremy, 1-800-955-6288 option 3, then option 1
- issue#: 405638599
They need to send to Indonesia for repair. 1 month eta.
20160519 update: tracking number for box (for us to put machine in and send to them)- 6506 8693 8148
Also:
*Hi Andrew,
You should receive a Loaner within 2-3 business days.
Thanks,
Foi Taua
Didn't know we were getting a loaner. He didn't mention cost.
20160520 update: Machine sent out
Page 8: 2016-05-18.Phylogenetic results without pogo sample
The results of phylogenetic analysis of SNP matrix from Page 3: 2016-05-16. Complete ddRAD-seq samples: processing. I excluded pogos, and it still needs further parsing.
- Get rid of LPR4, BSK. LPR4 is not in the right place. Also there was a labeling problem with this sample. BSK, have no clue what this sample actualyl is. It also had a labeling problem. BSK does not match any sites, but had KIte on the side. It is in the right place, but still have no clue which kite colony.
- Parse out bootstraps below 100
- Need to relabel kite samples so that they're lower case.
- Add in samples:
HW6-rudis
LPR4-ashmeadi
- 09A and 10A-rudis
Getting rid of LPR4 and BSK5
library(ape)x<-read.tree("20160518_ML_tree_BL_BS_RAxML.newick")plot(x)length(x$tip.label)x2<-drop.tip(x,c("LPR4","BSK5"))length(x2$tip.label)# checking lengthplot(x2) # plot to seewrite.tree(x2,"20160518_ML_tree_BL_BS_RAxML_parsed.newick") # new file nameParsing out bootstraps below 100
x2$node.label<-as.numeric(as.character(x2$node.label))x2$node.label<-ifelse(x2$node.label> 90,x2$node.label,"")x2$node.label[1]<-""x2$node.labelI'll hold off on adding samples to a phylogeny.
Transformed BL tree with 90 BS cutoff

Untransformed BL tree with 90 BS cutoff

Summary: Same topology without pogo sample.
Page 9: 2016-05-18. Agarose gel electrophoresis of qPCR amplicons; Curtis and ANBE samples
We wanted to check for specificity on a gel. Although, agarose gels don't completely pick up primer dimers. Even so, we acquire fluorescence at a higher temperature where those primer dimers disappear.
Sample list
| Lane | Section | Sample | Gene | Primer_pair |
|---|---|---|---|---|
| 1 | Top | Ladder | ||
| 2 | Top | Exit65 | hsc70-4 h2 | 1468+1592 |
| 3 | Top | BK | hsc70-4 h2 | 1468+1592 |
| 4 | Top | Ted6 | hsc70-4 h2 | 1468+1592 |
| 5 | Top | DUKE6 | hsc70-4 h2 | 1468+1592 |
| 6 | Top | ALA1 | hsc70-4 h2 | 1468+1592 |
| 7 | Top | KH2 | hsc70-4 h2 | 1468+1592 |
| 8 | Top | FB2 | hsc70-4 h2 | 1468+1592 |
| 9 | Top | Exit65 | hsp83 | 1592+1682 |
| 10 | Top | BK | hsp83 | 1592+1682 |
| 11 | Top | TED3 | hsp83 | 1592+1682 |
| 12 | Top | DUKE6 | hsp83 | 1592+1682 |
| 13 | Top | ALA1 | hsp83 | 1592+1682 |
| 14 | Top | KH2 | hsp83 | 1592+1682 |
| 15 | Top | FB2 | hsp83 | 1592+1682 |
| 16 | Top | PB1710 | hsp83 | 279 |
| 17 | Top | POP2 | hsp83 | 279 |
| 18 | Top | SHC2 | hsp83 | 279 |
| 19 | Top | cremato | hsp83 | 279 |
| 20 | Top | Ladder | ||
| 1 | Bottom | Ladder | ||
| 2 | Bottom | ex | hsp83 | 279 |
| 3 | Bottom | bk | hsp83 | 279 |
| 4 | Bottom | TED6 | hsp83 | 279 |
| 5 | Bottom | DUKE6 | hsp40 | 541+641 |
| 6 | Bottom | ALA1 | hsp40 | 541+641 |
| 7 | Bottom | KH2 | hsp40 | 541+641 |
| 8 | Bottom | FB2 | hsp40 | 541+641 |
| 9 | Bottom | EX | hsp40 | 541+641 |
| 10 | Bottom | BK | hsp40 | 541+641 |
| 11 | Bottom | Ted6 | hsp40 | 541+641 |
| 12 | Bottom | HF | hsp83 | 1592+1682 |
| 13 | Bottom | HF | hsp83 | 1592+1682 |
| 14 | Bottom | DF | hsp83 | 1592+1682 |
| 15 | Bottom | DF | hsp83 | 1592+1682 |
| 16 | Bottom | HF | hsc70-4 h2 | 1468+1592 |
| 17 | Bottom | HF | hsc70-4 h2 | 1468+1592 |
| 18 | Bottom | DF | hsc70-4 h2 | 1468+1592 |
| 19 | Bottom | DF | hsc70-4 h2 | 1468+1592 |
| 20 | Bottom | DF | actin | |
| 21 | Bottom | DF | actin | |
| 22 | Bottom | HF | hsp40 | 541+641 |
| 23 | Bottom | HF | hsp40 | 541+641 |
| 24 | Bottom | DF | hsp40 | 541+641 |
| 25 | Bottom | Ladder |
Protocol:
- Mixed ladder: 6.5 dye (6x) + 8 uL 100bp ladder+ 25.5 ul h20 to make 40 uL total---makes 4 lanes worth at 10 uL each lane
- For ANBE add 10 uL qpcr amplicon with 2 uL 6 x dye.
- For Curtis, add 5 uL qpcr amplicon with 1 uL 6 x dye.
- Electrophoresed on 1.5 % agarose gel , 125 Volts for 45 minutes.
Grayscaled whole:

Black whole:

The bottom is hard to see:
Showing pictures that focus on bottom part
Grayscaled bottom:

Black bottom:

Summary: Amplicons are specific. NO double bands.
Page 10: 2016-05-18. RaxML ML pairwise distance matrix
- Used SNP matrix found here: Page 3: 2016-05-16
- Used input tree(pre-parsed) found here: Page 8: 2016-05-18
Code for RaxML
./raxmlHPC -f x -p 12345 -s ~/Desktop/2015ANBE_common_garden/20160516Andrew_SNP_sequences.fas -m GTRGAMMA -t ~/Desktop/2015ANBE_common_garden/20160518ML_tree_unparsed.newick -n 20150618_ML_pairwise_distance_ANBEsamples
Snippet of output:
| V1 | V2 | V3 |
|---|---|---|
| HW5 | ALA1 | 0.094440 |
| HW5 | BK6-1 | 0.512869 |
| HW5 | POP3 | 0.096510 |
| HW5 | MA | 0.092071 |
| HW5 | CJ1 | 0.277364 |
| HW5 | Camb31-1 | 0.096856 |
| HW5 | DUKE9 | 0.113134 |
| HW5 | ALA2 | 0.098850 |
| HW5 | KH4 | 0.032412 |
| HW5 | Unit22-1 | 0.097533 |
Page 11: 2016-05-18. ABI steponeplus machine maintenance update
Update from, Page 7: 2016-05-17. ABI steponeplus machine maintenance.
*Hi Andrew,
Thank you for your recent request to have your StepOne Plus serial number 272007769 sent in to our Global Repair Center. Attached you will find the necessary paperwork to ensure that your unit is received correctly and promptly.
- Your RMA is 405638599
- Please review and complete the attached decontamination form, and print out 2 copies.
For 9700/9800's, Please put both the TOP and BASE serial numbers on the decontamination certificate. - Please DO NOT include your power cord with your instrument (remove from unit and keep it).
- Please DO NOT include any consumables (trays, tubes, etc.).
- Place a copy of the completed decontamination form INSIDE and OUTSIDE of the box.
- Print out the FedEx label, (link will arrive via separate email).
Service of your instrument cannot begin without the completed decontamination form.
Best Regards,
Foi Taua
Remote Service Center
T 800 955 6288 option 3, 1 to reach Remote Service Center
F 760 930 2300
5791 Van Allen Way • Carlsbad • CA • 92008 • United States
instrumentservices@lifetech.com
www.lifetechnologies.com*
2016-05-26 update: we received loaner.
Page 12: 2016-05-19. Getting whole rad loci with pyRAD tutorial and/or stacks
Previous analyses concatenate SNPS, but many studies use whole rad loci.
Computer cluster:
Reference for mason cluster
path of raw ddrad data
/N/dc2/scratch/scahan/Andrew_RADseq_051516/ Data/
SHC email:
*If you want to explore/analyze the RADseq data yourself:
/N/dc2/scratch/scahan/Aphaenogaster_RADfiles_051516/
You should find in each lane directory the raw .fq file from the sequencer, a barcode key file, the demultiplexed sample .fq files, and the trimming, filtering and mapping files from the pipeline. The earliest lanes (1&2) might have fewer files because the process was not yet regularized back then. The STACKs portion of the pipeline is specific to each project, so they all have their own directories in the main scratch space (e.g., Andrew_RADseq051516, Bernice051516, Phytotron_analyses_051516, etc.). All directories at this level have their date suffix modified every two weeks, so job scripts that point to a particular path have to get edited to the current date suffix. Some of the ddRAD lane directories also have a date suffix because they were secondarily moved from the main level into the Aphaenogaster directory.*
Trying pyRAD tutorial. Looks "easy".
No access to dependencies:
- scipy
- vsearch
- muscle
20160520 update, working on Mason compute cluster:
Hi Andrew,
First, I'd suggest you add "module load python" to your ~/.modules file, which will load the python 2.7.3 module each time you login. It's not terribly current, but it is the version under which we install python packages on Mason.
You'll find that numpy and scipy are both available there.
As for muscle and vsearch, I'll let you know when we get those packages installed.
Matt
I could use the population function/module in stacks.
Page 13: 2016-05-20. Evolution of proteome stability project
We are interested in the adaptive variation in how proteins unfold between 2 different ant species. Github repo
We isolated native proteins, subjected them to temperature treatments for 10 min. Then ultracentrifuged to pull down aggregates, then quantified. protocols here
Figure:

Function I am fitting to these points:
[Math Processing Error]

Code for curve fitting, also loading libraries
library(plyr)library(ggplot2)library(tidyr)library(minpack.lm)nls.fit<-function(data=data){ y<-nlsLM(unfolding ~ min+ (1-min)/(1+exp((-slope*(Tm-T)))),data=data, start=list(slope=.5,Tm=45,min=.3), trace=TRUE,control=nls.control(warnOnly = TRUE, tol = 1e-05, maxiter=1000)) #return(y) return(summary(y)$coefficients) }function to visualize curves by simply putting in paramters
fud<-function(T=seq(25,50,1),Tm=40,slope=.5,max=1,min=0){ y<-min+ (max-min)/(1+exp((-slope*(Tm-T)))) return(y) }How I implemented th code:
mod1<-ddply(x.par,.(Species,Colony),nls.fit)mod1$parameter<-rep(c("slope","Tm","min"),length(mod1$Species)/3)knitr::kable(mod1)Table summary of results from fitting curves.
| Species | Colony | Estimate | Std. Error | t value | Pr(>|t|) | parameter |
|---|---|---|---|---|---|---|
| A. rudis | Duke 1 | 0.1606280 | 0.0206403 | 7.782238 | 0.0000276 | slope |
| A. rudis | Duke 1 | 47.2920297 | 0.9451544 | 50.036301 | 0.0000000 | Tm |
| A. rudis | Duke 1 | 0.3637620 | 0.0293990 | 12.373285 | 0.0000006 | min |
| A. rudis | Lex 13 | 0.1333902 | 0.0159832 | 8.345673 | 0.0000158 | slope |
| A. rudis | Lex 13 | 49.7593929 | 1.2760137 | 38.995972 | 0.0000000 | Tm |
| A. rudis | Lex 13 | 0.2161279 | 0.0451703 | 4.784737 | 0.0009947 | min |
| A. rudis | Yates 2 | 0.1573466 | 0.0220329 | 7.141430 | 0.0000542 | slope |
| A. rudis | Yates 2 | 47.9849648 | 1.0899761 | 44.023870 | 0.0000000 | Tm |
| A. rudis | Yates 2 | 0.3637813 | 0.0336777 | 10.801853 | 0.0000019 | min |
| P. barbatus | WWRQ-45 | 0.2142567 | 0.0165774 | 12.924625 | 0.0000004 | slope |
| P. barbatus | WWRQ-45 | 45.9987927 | 0.3837543 | 119.865208 | 0.0000000 | Tm |
| P. barbatus | WWRQ-45 | 0.4032438 | 0.0126671 | 31.834069 | 0.0000000 | min |
| P. barbatus | WWRQ-53 | 0.1823480 | 0.0173963 | 10.482009 | 0.0000024 | slope |
| P. barbatus | WWRQ-53 | 47.2858982 | 0.5958843 | 79.354167 | 0.0000000 | Tm |
| P. barbatus | WWRQ-53 | 0.4013122 | 0.0184886 | 21.705927 | 0.0000000 | min |
| P. barbatus | WWRQ-8 | 0.2028211 | 0.0245990 | 8.245113 | 0.0000174 | slope |
| P. barbatus | WWRQ-8 | 45.5664742 | 0.6340253 | 71.868543 | 0.0000000 | Tm |
| P. barbatus | WWRQ-8 | 0.4280916 | 0.0194756 | 21.980921 | 0.0000000 | min |
Only slope was significant
summary(aov(Estimate~Species,data=slope)) Df Sum Sq Mean Sq F value Pr(>F) Species 1 0.003654 0.003654 15.15 0.0177 *Residuals 4 0.000965 0.000241 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Figure of unfolding if only changing slope (eye balled mean slope, so pogo = .2,rudis=.15 )

Page 14: 2016-05-24. Evolution of proteome stability project: Polyacrylamide gels for colony level replicates (A. rudis vs P. barbatus)
Amanda Meyer is working on this project:
- Samples stored at -80C, we took 30 uL and speed vacuumed (took 1 hr) and then resuspended in 60 uL of 1X sample buffer
- We took 25 uL of sample and added in 5 ul of gapdh (20ng/uL in sample buffer)
- Loaded on polyacryladmide and electrophoresed.
Polyacrylamide Gels:
- Duke1 (A. rudis)

- Yates2 (A. rudis)

Note: For Yates2, the gels are reversed. (Bottom gel starts at 30C) - WWRQ45 (P. barbatus)

- WWRQ53 (P. barbatus)

Next steps:
Need to destain and trypsin digest.
Page 15: 2016-05-24. Degenerate Hsp primer design from 2015-05-28
Primer design from 2015-05-28 referenced here
| n | name | sequence |
|---|---|---|
| 1 | hsc70-4h2_1175F | TTCTGYTGGAYGTDACTCC |
| 2 | hsc70-h2_1345R | TCGCTCTCTCHCCYTCRTARAC |
| 3 | hsc70-4h2_1468F | GCGATYGARAAATCTACVGGC |
| 4 | hsc7004h2_1592R | TGYTCRTCYTCCGATCGGTA |
| 5 | hsc70-4h1_1291F | ACYTAYGCCGACAATCARCC |
| 6 | hsc70-4h2_1390R | CGCTRAGCTCGAAYTTDCCC |
| 7 | hsc70-4h1_1506F | CACYATYACCAAYGACAARG |
| 8 | hsc70-4h1_1605R | YTCCTTCTGCTTCTCRTCCTC |
| 9 | hsp40_118F | GCCTTRCGATATCATCCTGA |
| 10 | hsp40_248 | CCYTCCTCGCCRAATTTATC |
| 11 | hsp40_541F | AAAGATCGYGCYCARGATCC |
| 12 | hsp40_641R | GCYCGTCTRCATATYTTCATC |
| 13 | hsp40_869F | TRTGCGGTACTRTYGTCGAAG |
| 14 | hsp40_968R | TGGAACCTYTTGACNGTRTTC |
| 15 | hsp83_278F | ACDATYCTTGATTCTGGYATTGG |
| 16 | hsp83_392R | CCAAACTGTCCAATCATGGA |
| 17 | hsp83754F | GATGTYGGHGAGGATGA |
| 18 | hsp83_880R | GATTTCTYGTCCARATCGG |
| 19 | hsp83_1583F | AATTCGAYGGAAARCAGYTGG |
| 20 | hsp83_1682R | AAYTTGGCYTTGTCYTCCTC |
| 21 | hsp83_1807F | ATGGAGAGRATCATGAAGGC |
| 22 | hsp83_1917R | CARRTTCTCCATGATRGGATGATC |
| 23 | nedd_510F | TAATCATTCCAGTCAGCGG |
| 24 | ned_614R | TCAGATACGTCTCCGTTGTC |
| 25 | nedd_556F | TATCATGCATACATTTCCGAC |
| 26 | nedd_683R | ATCGTAATATCTGCACTTTGYTC |
| 27 | nedd_956F | ATGGTGAAGTTCTACGCGAG |
| 28 | nedd_1088R | TAAGGTAGCCACGTTGATCG |
| 29 | nedd_1222F | CAAGTAGCACCTAATGGTAGA |
| 30 | nedd_1316R | GGTATAGARCTTGGTCTTCC |
| 31 | nedd_1351F | GATTTAGATCAATTAGGACCDCTTC |
| 32 | nedd_1460R | GGATCTTCCCATTGTGTTGT |
| 33 | nedd_2375F | GGAGAGTCGTTTTGTCATTCAG |
| 34 | nedd_2459R | CCATTCATTGGAACACGTGATG |
I don't use all of these anymore. But here are the ones that I've tested for specificity (from agarose gel electrophoresis , sequencing, and melt curve analysis following qPCR) and efficiency ( titrate amplicon across a dynamic range to compare slope equals -3.2).
- hsc70-4 h2; 1468F + 1592R
- hsp83; 278F+392R and 1583F + 1682 R
- hsp40 541F+ 641R
- NEDD; 956F+ 1088R (This is off the top of my head, so I need to double check this!)
Page 16: 2016-05-24](#id-section16). Sequencing analysis continued from Page 5: 2016-05-16.
Sharing screenshots of sanger sequenced samples mapped to reference transcript (P. barbatus)
I used the software Geneious v6 to analyze sequence data.
Sample structure on figure: Well_colony.id_gene_primer#
The pics and raw sequence data can be found: here
- Path: /Dissertation_temperature_adaptation_ants/Dissertation_Projects/2014_xanbe-common-garden_gxp_evolution/Data/sequencing/Sanger/
1. hsc70-4 h2 1468F + 1592R

2. hsp83 278F+392R

3. hsp83 1583F + 1682 R

4. hsp40 541F+ 641R

Summary of results:
Most of samples mapped really well! Generally, the sequencing with the forward primer recovers the reverse primer, and vice versa.
Page 17: 2016-05-25. Double check samples for SHC; JSG phytotron exp and MS.
email sent 2016-05-18:
Ok – your list is missing 20-B (AP2), which is on your tree. The two samples with no morphological ID are your RW2 (25-C) and your BP2 (07-B), which will have to get omitted. The only remaining samples whose placements are problematic are your RW1, which was ID’d picea but comes out in that odd basal clade with the intermediate NK samples, and LA4, which was ID’d as rudis but falls out in the middle of picea. Looking at the latter one, however, this is the mysterious LVA9, which was written down as the source for two different experimental colonies (not possible, since they were supposed to be queenright) and there is no way to know if the sample Bernice looked at is the same as the RADseq sample or the assayed colony. So there is good reason to throw that one out as well.
Need to double check these samples.
2016-05-26 UPdate- Ecluding:
- 25-C / RW2
- 07-B / BP2
- 10-F / LA4
Page 18: 2016-05-31. Learning model selection and model averaging!
I'm learning model averaging!
Basically, there is uncertainty in parameter estimates of a stat model (linear regression) and we should explore how many stats model compare to each other, usually by AIC.
From Burnham and Anderson 2002
If data analysis relies on model selection, then inferences should acknowledge model selection uncertainty. If the goal is to get the best estimates of a set of parameters in common to all models (this includes prediction), model averaging is recommended. If the models have definite, and differing, interpretations as regards understanding relationships among variables, and it is such understanding that is sought, then one wants to identify the best model and make inferences based on that model. Hence, reported parameter estimates should then be from the selected model (not model averaged values). However, even when selecting a best model, also note the competing models, as ranked by their Akaike weights. Restricting detailed comparisons to the models in a 90% confidence set on models should often suffice. If a single model is not strongly supported, wmin ≥ 0.9, and competing models give alternative inferences, this should be reported. It may occur that the basic inference(s) will be the same from all good models. However, this is not always the case, and then inference based on a single best model may not be sound if support for even the best model is weak (in all-subsets selection when R > 1,000, wmin can be verysmall, e.g., < 0.01).
General Steps:
- Construct global model. Pick predictors you think are most important.
- I used MuMin package in R with the dredge() function to construct subsets of global model.
- Pick out top model set: subset based on.... top 2/6/10 AIC or delta 4 AIC.
- Average models from top set.
Picking predictors I think are important
Decomposing phylogeny with PCOA, looking at eigenvalues:
| Eigenvalues | Relative_eig | Rel_corr_eig | Broken_stick | Cum_corr_eig | Cumul_br_stick | rep(1:20, 1) |
|---|---|---|---|---|---|---|
| 0.362 | 0.563 | 0.407 | 0.114 | 0.407 | 0.114 | 1 |
| 0.086 | 0.134 | 0.102 | 0.087 | 0.509 | 0.200 | 2 |
| 0.052 | 0.081 | 0.065 | 0.073 | 0.574 | 0.273 | 3 |
| 0.020 | 0.032 | 0.030 | 0.064 | 0.604 | 0.337 | 4 |
| 0.016 | 0.025 | 0.025 | 0.057 | 0.630 | 0.394 | 5 |
| 0.014 | 0.022 | 0.023 | 0.052 | 0.653 | 0.446 | 6 |
| 0.011 | 0.017 | 0.020 | 0.047 | 0.673 | 0.494 | 7 |
| 0.010 | 0.015 | 0.018 | 0.043 | 0.691 | 0.537 | 8 |
| 0.008 | 0.013 | 0.017 | 0.040 | 0.708 | 0.577 | 9 |
| 0.007 | 0.011 | 0.016 | 0.037 | 0.723 | 0.614 | 10 |
| 0.005 | 0.008 | 0.013 | 0.034 | 0.737 | 0.649 | 11 |
| 0.005 | 0.008 | 0.013 | 0.032 | 0.750 | 0.681 | 12 |
| 0.004 | 0.007 | 0.013 | 0.030 | 0.762 | 0.710 | 13 |
| 0.004 | 0.007 | 0.012 | 0.028 | 0.775 | 0.738 | 14 |
| 0.004 | 0.006 | 0.012 | 0.026 | 0.787 | 0.764 | 15 |
| 0.004 | 0.006 | 0.012 | 0.024 | 0.798 | 0.787 | 16 |
| 0.003 | 0.005 | 0.011 | 0.022 | 0.810 | 0.810 | 17 |
| 0.003 | 0.005 | 0.011 | 0.021 | 0.821 | 0.830 | 18 |
| 0.003 | 0.005 | 0.011 | 0.019 | 0.832 | 0.849 | 19 |
| 0.003 | 0.005 | 0.011 | 0.018 | 0.843 | 0.867 | 20 |
We have ~40 samples, so use 10:1 rule (sample: predictor). Regress first 4 Axes (60% of variation) against Ctmax.
Ctmax.sel<-lm(Ctmax~Axis.1+Axis.2+Axis.3+Axis.4,data=merg)summary(Ctmax.sel) Estimate Std. Error t value Pr(>|t|) (Intercept) 42.43692 0.06339 669.409 <2e-16 ***Axis.1 11.87677 0.65817 18.045 <2e-16 ***Axis.2 2.87094 1.35038 2.126 0.0408 * Axis.3 3.72343 1.73540 2.146 0.0391 * Axis.4 -2.25911 2.76538 -0.817 0.4197 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3959 on 34 degrees of freedomMultiple R-squared: 0.908, Adjusted R-squared: 0.8971 F-statistic: 83.85 on 4 and 34 DF, p-value: < 2.2e-16Looksl ike first 3 axes are significant: choose these in regression models with Tmax
Check correlation between bioclim variables and phylogenetic components
| Axis.1 | Axis.2 | Axis.3 | Axis.4 | bio5 | bio6 | bio7 | merg$nb | |
|---|---|---|---|---|---|---|---|---|
| Axis.1 | 1.000 | 0.000 | 0.000 | 0.000 | 0.882 | 0.745 | -0.454 | -0.258 |
| Axis.2 | 0.000 | 1.000 | 0.000 | 0.000 | 0.159 | 0.139 | -0.089 | 0.023 |
| Axis.3 | 0.000 | 0.000 | 1.000 | 0.000 | 0.151 | 0.301 | -0.327 | -0.321 |
| Axis.4 | 0.000 | 0.000 | 0.000 | 1.000 | -0.044 | -0.090 | 0.099 | 0.072 |
| bio5 | 0.882 | 0.159 | 0.151 | -0.044 | 1.000 | 0.772 | -0.411 | -0.412 |
| bio6 | 0.745 | 0.139 | 0.301 | -0.090 | 0.772 | 1.000 | -0.897 | -0.728 |
| bio7 | -0.454 | -0.089 | -0.327 | 0.099 | -0.411 | -0.897 | 1.000 | 0.757 |
| merg$nb | -0.258 | 0.023 | -0.321 | 0.072 | -0.412 | -0.728 | 0.757 | 1.000 |
Model subsets
Construct full model to test interaction between Tma and each eigenvector(part of phylogeny
#Ctmax = upper thermal limit# Axis1 - picea rudis split# Axis2 - N-S rudis clade split# Axis 3 - Pica split# Rearing temp: 20(23?) and 26#Bio 5 = Tmaxlm(Ctmax~bio5*Axis.1+bio5*Axis.2+bio5*Axis.3+Rearing.temp,data=merg) Showing table of model subsets generated from dredge() function
| (Intercept) | Axis.1 | Axis.2 | Axis.3 | bio5 | Rearing.temp | Axis.1:bio5 | Axis.2:bio5 | Axis.3:bio5 | df | logLik | AICc | delta | weight | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 112 | 38.89633 | -64.54042 | 163.379439 | -7.1344145 | 0.1142023 | NA | 2.536627 | -5.254042 | NA | 8 | -7.240543 | 35.28109 | 0.000000 | 0.4265037 |
| 42 | 40.27150 | -32.80201 | NA | NA | 0.0688105 | NA | 1.425184 | NA | NA | 5 | -13.003298 | 37.82478 | 2.543691 | 0.1195549 |
| 128 | 38.47028 | -66.64409 | 164.835542 | -7.4660612 | 0.1213695 | 0.0095824 | 2.601996 | -5.309316 | NA | 9 | -7.070997 | 38.34889 | 3.067805 | 0.0919936 |
| 240 | 38.99929 | -63.44634 | 165.175057 | -17.1348640 | 0.1105860 | NA | 2.500507 | -5.300206 | 0.3704068 | 9 | -7.230877 | 38.66865 | 3.387565 | 0.0784011 |
| 44 | 39.58611 | -45.63541 | -2.113326 | NA | 0.0908785 | NA | 1.833044 | NA | NA | 6 | -12.197967 | 39.02093 | 3.739847 | 0.0657393 |
| 108 | 40.67312 | -34.07965 | 68.733204 | NA | 0.0549570 | NA | 1.516668 | -2.192978 | NA | 7 | -10.725474 | 39.06385 | 3.782765 | 0.0643437 |
| 46 | 40.41404 | -31.25480 | NA | 0.5303359 | 0.0639742 | NA | 1.377435 | NA | NA | 6 | -12.955310 | 40.53562 | 5.254534 | 0.0308259 |
| 58 | 40.27478 | -32.80931 | NA | NA | 0.0687900 | -0.0001219 | 1.425440 | NA | NA | 6 | -13.003275 | 40.63155 | 5.350465 | 0.0293822 |
| 48 | 39.02228 | -53.61002 | -2.839872 | -1.2211435 | 0.1096013 | NA | 2.083210 | NA | NA | 7 | -12.029592 | 41.67209 | 6.391001 | 0.0174636 |
| 60 | 39.47928 | -45.71692 | -2.160369 | NA | 0.0919412 | 0.0034085 | 1.834965 | NA | NA | 7 | -12.180302 | 41.97351 | 6.692423 | 0.0150204 |
| 256 | 38.58207 | -65.43836 | 166.850595 | -18.6312806 | 0.1173856 | 0.0096530 | 2.562160 | -5.361253 | 0.4134582 | 10 | -7.058857 | 41.97486 | 6.693772 | 0.0150103 |
| 124 | 40.68974 | -34.04638 | 68.875613 | NA | 0.0547434 | -0.0004636 | 1.515799 | -2.197188 | NA | 8 | -10.725127 | 42.25025 | 6.969168 | 0.0130794 |
| 174 | 40.17971 | -36.89376 | NA | -39.0161170 | 0.0707023 | NA | 1.545609 | NA | 1.4424428 | 7 | -12.647902 | 42.90871 | 7.627622 | 0.0094104 |
| 62 | 40.43434 | -31.27748 | NA | 0.5367575 | 0.0637997 | -0.0006914 | 1.378308 | NA | NA | 7 | -12.954602 | 43.52211 | 8.241021 | 0.0069248 |
| 176 | 38.62972 | -58.09940 | -4.104072 | 36.3065961 | 0.1233954 | NA | 2.234488 | NA | -1.3972498 | 8 | -11.917041 | 44.63408 | 9.352996 | 0.0039714 |
| 64 | 38.74224 | -54.92127 | -3.032774 | -1.3987999 | 0.1142952 | 0.0063182 | 2.123167 | NA | NA | 8 | -11.971892 | 44.74378 | 9.462699 | 0.0037594 |
| 76 | 42.07767 | 13.63416 | 119.882620 | NA | 0.0151404 | NA | NA | -3.677760 | NA | 6 | -15.148179 | 44.92136 | 9.640273 | 0.0034400 |
| 8 | 42.43692 | 11.87677 | 2.870941 | 3.7234291 | NA | NA | NA | NA | NA | 5 | -16.905003 | 45.62819 | 10.347102 | 0.0024159 |
| 190 | 40.09493 | -37.03074 | NA | -40.5928212 | 0.0716161 | 0.0025740 | 1.548960 | NA | 1.4990805 | 8 | -12.638411 | 46.07682 | 10.795736 | 0.0019305 |
| 6 | 42.43692 | 11.87677 | NA | 3.7234291 | NA | NA | NA | NA | NA | 4 | -19.294824 | 47.76612 | 12.485033 | 0.0008295 |
Cumulative AIC weights

2016-06-01 continued : Actually model averaging
top 2 AIC
>summary(model.avg(a.max[1:2]))Full model-averaged coefficients (with shrinkage): Estimate Std. Error Adjusted SE z value Pr(>|z|) (Intercept) 39.19741 1.81718 1.88065 20.842 < 2e-16 ***Axis.1 -57.59157 22.32729 22.90080 2.515 0.01191 * Axis.2 127.60890 83.60797 84.76171 1.506 0.13220 Axis.3 -5.57240 3.87984 3.94483 1.413 0.15778 bio5 0.10426 0.06385 0.06611 1.577 0.11477 Axis.1:bio5 2.29329 0.74450 0.76259 3.007 0.00264 ** Axis.2:bio5 -4.10371 2.67227 2.70829 1.515 0.12971 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Relative variable importance: Axis.1 bio5 Axis.1:bio5 Axis.2 Axis.3 Axis.2:bio5Importance: 1.00 1.00 1.00 0.78 0.78 0.78 N containing models: 2 2 2 1 1 1 top 6 AIC
>summary(model.avg(a.max[1:6]))Full model-averaged coefficients (with shrinkage): Estimate Std. Error Adjusted SE z value Pr(>|z|) (Intercept) 39.242396 1.895875 1.960765 20.014 < 2e-16 ***Axis.1 -56.401968 22.642086 23.231252 2.428 0.01519 * Axis.2 120.584643 84.389355 85.517189 1.410 0.15852 Axis.3 -5.992746 25.128163 26.111410 0.230 0.81848 bio5 0.101921 0.065747 0.068039 1.498 0.13414 Axis.1:bio5 2.251255 0.751716 0.770321 2.922 0.00347 ** Axis.2:bio5 -3.881627 2.688644 2.723787 1.425 0.15413 Rearing.temp 0.001041 0.006764 0.006986 0.149 0.88151 Axis.3:bio5 0.034305 0.915567 0.952226 0.036 0.97126 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Relative variable importance: Axis.1 bio5 Axis.1:bio5 Axis.2 Axis.2:bio5 Axis.3Importance: 1.00 1.00 1.00 0.86 0.78 0.71 N containing models: 6 6 6 5 4 3 Rearing.temp Axis.3:bio5Importance: 0.11 0.09 N containing models: 1 1 top 10 AIC
>summary(model.avg(a.max[1:10])) Full model-averaged coefficients (with shrinkage): Estimate Std. Error Adjusted SE z value Pr(>|z|) (Intercept) 3.931e+01 1.901e+00 1.965e+00 20.004 < 2e-16 ***Axis.1 -5.462e+01 2.281e+01 2.337e+01 2.337 0.01945 * Axis.2 1.086e+02 8.793e+01 8.891e+01 1.221 0.22190 Axis.3 -5.407e+00 2.393e+01 2.486e+01 0.218 0.82782 bio5 9.962e-02 6.590e-02 6.817e-02 1.461 0.14391 Axis.1:bio5 2.187e+00 7.598e-01 7.775e-01 2.813 0.00491 ** Axis.2:bio5 -3.499e+00 2.803e+00 2.833e+00 1.235 0.21689 Rearing.temp 9.893e-04 7.724e-03 7.987e-03 0.124 0.90143 Axis.3:bio5 3.092e-02 8.693e-01 9.041e-01 0.034 0.97272 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Relative variable importance: Axis.1 bio5 Axis.1:bio5 Axis.2 Axis.2:bio5 Axis.3Importance: 1.00 1.00 1.00 0.81 0.70 0.69 N containing models: 10 10 10 7 4 5 Rearing.temp Axis.3:bio5Importance: 0.15 0.08 N containing models: 3 1 top 4 delta AIC
>summary(model.avg(a.max, subset = delta < 4))Full model-averaged coefficients (with shrinkage): Estimate Std. Error Adjusted SE z value Pr(>|z|) (Intercept) 39.242396 1.895875 1.960765 20.014 < 2e-16 ***Axis.1 -56.401968 22.642086 23.231252 2.428 0.01519 * Axis.2 120.584643 84.389355 85.517189 1.410 0.15852 Axis.3 -5.992746 25.128163 26.111410 0.230 0.81848 bio5 0.101921 0.065747 0.068039 1.498 0.13414 Axis.1:bio5 2.251255 0.751716 0.770321 2.922 0.00347 ** Axis.2:bio5 -3.881627 2.688644 2.723787 1.425 0.15413 Rearing.temp 0.001041 0.006764 0.006986 0.149 0.88151 Axis.3:bio5 0.034305 0.915567 0.952226 0.036 0.97126 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Relative variable importance: Axis.1 bio5 Axis.1:bio5 Axis.2 Axis.2:bio5 Axis.3Importance: 1.00 1.00 1.00 0.86 0.78 0.71 N containing models: 6 6 6 5 4 3 Rearing.temp Axis.3:bio5Importance: 0.11 0.09 N containing models: 1 1 Comparing output to stepwise AIC both directions
> summary(stepAIC(full.max,direction="both"))Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 38.89633 1.77085 21.965 < 2e-16 ***bio5 0.11420 0.06223 1.835 0.075805 . Axis.1 -64.54042 19.60370 -3.292 0.002429 ** Axis.2 163.37944 55.72789 2.932 0.006179 ** Axis.3 -7.13441 2.85109 -2.502 0.017640 * bio5:Axis.1 2.53663 0.63426 3.999 0.000351 ***bio5:Axis.2 -5.25404 1.76036 -2.985 0.005402 ** ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3216 on 32 degrees of freedomMultiple R-squared: 0.9428, Adjusted R-squared: 0.9321 F-statistic: 87.96 on 6 and 32 DF, p-value: < 2.2e-16SHC suggestion: Just include all phylo axes in all analyses
full.max<-lm(Ctmax~bio5*Axis.1+bio5*Axis.2+bio5*Axis.3+bio5*Axis.4+Rearing.temp,data=merg)Showing top 2 AIC
summary(model.avg(a.max[1:2]))Full model-averaged coefficients (with shrinkage): Estimate Std. Error Adjusted SE z value Pr(>|z|) (Intercept) 39.19741 1.81718 1.88065 20.842 < 2e-16 ***Axis.1 -57.59157 22.32729 22.90080 2.515 0.01191 * Axis.2 127.60890 83.60797 84.76171 1.506 0.13220 Axis.3 -5.57240 3.87984 3.94483 1.413 0.15778 bio5 0.10426 0.06385 0.06611 1.577 0.11477 Axis.1:bio5 2.29329 0.74450 0.76259 3.007 0.00264 ** Axis.2:bio5 -4.10371 2.67227 2.70829 1.515 0.12971 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Relative variable importance: Axis.1 bio5 Axis.1:bio5 Axis.2 Axis.3 Axis.2:bio5Importance: 1.00 1.00 1.00 0.78 0.78 0.78 N containing models: 2 2 2 1 1 1 Showing stepwise variable selection
> summary(stepAIC(full.max,direction="both"))Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 38.89633 1.77085 21.965 < 2e-16 ***bio5 0.11420 0.06223 1.835 0.075805 . Axis.1 -64.54042 19.60370 -3.292 0.002429 ** Axis.2 163.37944 55.72789 2.932 0.006179 ** Axis.3 -7.13441 2.85109 -2.502 0.017640 * bio5:Axis.1 2.53663 0.63426 3.999 0.000351 ***bio5:Axis.2 -5.25404 1.76036 -2.985 0.005402 ** ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3216 on 32 degrees of freedomMultiple R-squared: 0.9428, Adjusted R-squared: 0.9321 F-statistic: 87.96 on 6 and 32 DF, p-value: < 2.2e-162016-06-02 update:
full mod construction for all traits
#Ctmaxfull.max<-lm(Ctmax~bio5*Axis.1+bio5*Axis.2+bio5*Axis.3+bio5*Axis.4+Rearing.temp,data=merg)#Ctminfull.min<-lm(Ctmin~bio6*Axis.1+bio6*Axis.2+bio6*Axis.3+bio6*Axis.4+Rearing.temp,data=merg)#thermal tolerance breadthTNB.full<-lm(nb~Axis.1*bio7+Axis.2*bio7+Axis.3*bio7+Axis.4*bio7+Rearing.temp,data=merg)Probably a poor way to show output, but you can see the consistency with model averaging at different criteria for selecting top model (Top 2/6/10 AIC, < delta 4, 95 conf int):

full table
| Ctmax | X | X.1 | X.2 | X.3 | X.4 | X.5 | Ctmin | X.6 | X.7 | X.8 | X.9 | X.10 | X.11 | TNB | X.12 | X.13 | X.14 | X.15 | X.16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| top 2 AICc | NA | top 2 AICc | NA | top 2 AICc | |||||||||||||||
| Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | |||
| (Intercept) | 39.1974113 | 1.81717555 | 1.88065146 | 20.842464 | 0 | NA | (Intercept) | 6.7081182 | 0.2276948 | 0.23542884 | 28.4931876 | 0 | NA | (Intercept) | 27.0459363 | 1.9059241 | 1.96848068 | 13.7394979 | 0 |
| Axis.1 | -57.5915669 | 22.32728931 | 22.90079918 | 2.514828 | 0.01190905 | NA | Axis.2 | -2.1928922 | 2.3776364 | 2.41435204 | 0.9082736 | 0.3637337 | NA | bio7 | 0.3458702 | 0.05107519 | 0.05275118 | 6.5566338 | 0 |
| Axis.2 | 127.6089015 | 83.60796871 | 84.76171296 | 1.505502 | 0.13219514 | NA | bio6 | 0.4381219 | 0.0216451 | 0.02237847 | 19.5778289 | 0 | NA | Axis.1 | 0.4799895 | 1.20994269 | 1.23717296 | 0.3879728 | 0.6980362 |
| Axis.3 | -5.5723952 | 3.87984439 | 3.94482507 | 1.412584 | 0.1577782 | NA | NA | ||||||||||||
| bio5 | 0.1042642 | 0.06385464 | 0.06611108 | 1.577106 | 0.11477121 | NA | NA | ||||||||||||
| Axis.1:bio5 | 2.2932857 | 0.74450301 | 0.76259317 | 3.00722 | 0.00263649 | NA | NA | ||||||||||||
| Axis.2:bio5 | -4.1037142 | 2.67226625 | 2.70829115 | 1.515241 | 0.12971134 | NA | NA | ||||||||||||
| NA | NA | ||||||||||||||||||
| top 6 AICc | NA | top 6 AIC | NA | top 6 AIC | |||||||||||||||
| Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | |||
| (Intercept) | 39.09770833 | 1.858677254 | 1.925749929 | 20.30258848 | 0 | NA | (Intercept) | 6.684988297 | 0.34810913 | 0.35955592 | 18.59234664 | 0 | NA | (Intercept) | 27.10029979 | 1.93125923 | 1.9947008 | 13.58614773 | 0 |
| Axis.1 | -58.89921078 | 22.04271462 | 22.67258208 | 2.59781663 | 0.0093819 | NA | Axis.2 | -2.395005615 | 2.383652 | 2.42548151 | 0.98743511 | 0.3234294 | NA | bio7 | 0.340556895 | 0.05066679 | 0.05234842 | 6.505580624 | 0 |
| Axis.2 | 128.5516463 | 84.09957359 | 85.29260627 | 1.50718394 | 0.1317635 | NA | bio6 | 0.434621522 | 0.02580208 | 0.02660974 | 16.33317269 | 0 | NA | Axis.1 | 0.233929172 | 0.87808967 | 0.89639122 | 0.26096772 | 0.7941174 |
| Axis.3 | -6.557056311 | 24.86761943 | 25.84512303 | 0.25370575 | 0.7997229 | NA | Axis.1 | 0.237902792 | 0.84028327 | 0.86112014 | 0.27627131 | 0.7823397 | NA | Rearing.temp | 0.006336015 | 0.02480004 | 0.02533232 | 0.250115874 | 0.8024977 |
| bio5 | 0.106760957 | 0.064593101 | 0.066955014 | 1.59451773 | 0.1108201 | NA | Axis.4 | 0.112568203 | 1.35995056 | 1.40562657 | 0.080084 | 0.9361704 | NA | Axis.2 | 0.384777978 | 1.53920801 | 1.57274344 | 0.244654003 | 0.8067243 |
| Axis.1:bio5 | 2.335012045 | 0.732742988 | 0.752658592 | 3.10235221 | 0.0019199 | NA | Rearing.temp | -0.000455203 | 0.01026543 | 0.01062611 | 0.04283813 | 0.9658306 | NA | Axis.3 | -0.405958108 | 1.89199147 | 1.93749909 | 0.209526864 | 0.834037 |
| Axis.2:bio5 | -4.139188942 | 2.678608124 | 2.715902917 | 1.5240563 | 0.1274946 | NA | NA | Axis.4 | -0.020203078 | 2.08189989 | 2.15420639 | 0.009378432 | 0.9925172 | ||||||
| Rearing.temp | 0.001023202 | 0.006706439 | 0.006926472 | 0.14772334 | 0.8825611 | NA | NA | ||||||||||||
| Axis.4 | 0.040415014 | 0.730962899 | 0.759753398 | 0.05319491 | 0.9575766 | NA | NA | ||||||||||||
| Axis.3:bio5 | 0.033707897 | 0.907576414 | 0.943914621 | 0.03571075 | 0.971513 | NA | NA | ||||||||||||
| NA | NA | ||||||||||||||||||
| top10 AICc | NA | top 10 AIC | NA | top 10 AIC | |||||||||||||||
| Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | |||
| (Intercept) | 3.93E+01 | 1.899791736 | 1.963911039 | 20.01693332 | 0 | NA | (Intercept) | 6.708401068 | 0.37935474 | 0.39165216 | 17.1284671 | 0 | NA | (Intercept) | 26.99566918 | 1.97438106 | 2.03890627 | 13.2402698 | 0 |
| Axis.1 | -5.49E+01 | 22.98798186 | 23.54781004 | 2.33039265 | 0.0197854 | NA | Axis.2 | -2.306237463 | 2.41991077 | 2.46193139 | 0.936759437 | 0.3488823 | NA | bio7 | 0.341349078 | 0.05167576 | 0.05337701 | 6.3950576 | 0 |
| Axis.2 | 1.13E+02 | 87.18011426 | 88.20794399 | 1.28066363 | 0.2003118 | NA | bio6 | 0.434528345 | 0.0260478 | 0.0268662 | 16.17379177 | 0 | NA | Axis.1 | 0.333219277 | 1.03492288 | 1.0576099 | 0.3150682 | 0.7527099 |
| Axis.3 | -5.52E+00 | 22.98845903 | 23.88232484 | 0.23133093 | 0.8170577 | NA | Axis.1 | 0.134917912 | 0.91388635 | 0.93807943 | 0.143823548 | 0.8856398 | NA | Rearing.temp | 0.009351372 | 0.0297777 | 0.03043434 | 0.3072639 | 0.7586425 |
| bio5 | 9.98E-02 | 0.06595076 | 0.068221677 | 1.46265473 | 0.1435619 | NA | Axis.4 | 0.084008834 | 1.17585796 | 1.21528353 | 0.069126942 | 0.9448886 | NA | Axis.2 | 0.449552864 | 1.65384841 | 1.69015606 | 0.265983 | 0.7902523 |
| Axis.1:bio5 | 2.20E+00 | 0.766278694 | 0.783913018 | 2.80383221 | 0.0050499 | NA | Rearing.temp | -0.001159531 | 0.01228646 | 0.01268573 | 0.091404373 | 0.9271713 | NA | Axis.3 | -0.513063577 | 2.10220292 | 2.15127673 | 0.2384926 | 0.8114991 |
| Axis.2:bio5 | -3.64E+00 | 2.782129711 | 2.81413218 | 1.29206427 | 0.1963349 | NA | Axis.3 | -0.018400092 | 0.73655574 | 0.76270452 | 0.024124797 | 0.9807531 | NA | Axis.4 | -32.1405597 | 171.6554339 | 174.0144258 | 0.1847005 | 0.8534639 |
| Rearing.temp | 8.61E-04 | 0.007016252 | 0.007252224 | 0.1187358 | 0.9054847 | NA | Axis.2:bio6 | -0.001287445 | 0.19407722 | 0.20101898 | 0.006404594 | 0.9948899 | NA | Axis.4:bio7 | 0.985550403 | 5.26653 | 5.33891153 | 0.1845976 | 0.8535446 |
| Axis.4 | -5.58E-03 | 0.843672767 | 0.874007493 | 0.00638567 | 0.994905 | NA | Axis.1:bio6 | -0.02598336 | 0.13293313 | 0.13465174 | 0.192967133 | 0.8469847 | NA | ||||||
| Axis.3:bio5 | 2.85E-02 | 0.834371842 | 0.867772028 | 0.03282359 | 0.9738153 | NA | NA | ||||||||||||
| NA | NA | ||||||||||||||||||
| delta 4 | NA | delta 4 | NA | delta 4 | |||||||||||||||
| Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | |||
| (Intercept) | 3.92E+01 | 1.893976934 | 1.959535391 | 20.00841235 | 0 | NA | (Intercept) | 6.705813971 | 0.36859922 | 0.38057547 | 17.62019506 | 0 | NA | (Intercept) | 26.99362954 | 1.98319436 | 2.04854756 | 13.1769601 | 0 |
| Axis.1 | -5.72E+01 | 22.60477016 | 23.20900583 | 2.46345624 | 0.0137605 | NA | Axis.2 | -2.092714799 | 2.40013254 | 2.43860238 | 0.85816155 | 0.3908033 | NA | bio7 | 0.341410566 | 0.05185096 | 0.05357386 | 6.3727077 | 0 |
| Axis.2 | 1.24E+02 | 83.35210021 | 84.53449839 | 1.4715241 | 0.1411494 | NA | bio6 | 0.434592864 | 0.02574957 | 0.0265647 | 16.35978856 | 0 | NA | Axis.1 | 0.332214471 | 1.03734215 | 1.06050509 | 0.3132606 | 0.7540827 |
| Axis.3 | -6.10E+00 | 24.04585344 | 24.98659536 | 0.24418582 | 0.8070869 | NA | Axis.1 | 0.122426556 | 0.87143086 | 0.89445417 | 0.136872923 | 0.8911312 | NA | Rearing.temp | 0.009346593 | 0.02990764 | 0.03058437 | 0.3056003 | 0.759909 |
| bio5 | 1.03E-01 | 0.065747663 | 0.068062898 | 1.51566905 | 0.1296031 | NA | Axis.4 | 0.125415028 | 1.4735481 | 1.52274366 | 0.082361222 | 0.9343595 | NA | Axis.2 | 2.838029586 | 17.73196229 | 18.10766781 | 0.1567308 | 0.875457 |
| Axis.1:bio5 | 2.28E+00 | 0.749621892 | 0.768722868 | 2.96354009 | 0.0030412 | NA | Rearing.temp | -0.001052176 | 0.0117087 | 0.01208889 | 0.087036633 | 0.9306424 | NA | Axis.3 | -0.629285835 | 2.34142278 | 2.39958368 | 0.2622479 | 0.7931303 |
| Axis.2:bio5 | -4.00E+00 | 2.655253834 | 2.692134467 | 1.48727215 | 0.1369429 | NA | Axis.3 | -0.018674382 | 0.92571971 | 0.95829077 | 0.019487177 | 0.9844525 | NA | Axis.4 | -27.62992928 | 159.5462112 | 161.7281284 | 0.1708418 | 0.8643481 |
| Rearing.temp | 9.52E-04 | 0.006474441 | 0.006686524 | 0.14238996 | 0.886772 | NA | Axis.2:bio6 | -0.001168247 | 0.18487511 | 0.19148771 | 0.006100898 | 0.9951322 | NA | Axis.4:bio7 | 0.847237516 | 4.89499575 | 4.96194424 | 0.1707471 | 0.8644226 |
| Axis.4 | 3.76E-02 | 0.705181274 | 0.732950521 | 0.05130819 | 0.9590799 | NA | Axis.1:bio6 | -0.023577694 | 0.12685366 | 0.12848792 | 0.183501249 | 0.8544047 | NA | Axis.2:bio7 | -0.064576645 | 0.51493977 | 0.52615353 | 0.1227335 | 0.9023182 |
| Axis.3:bio5 | 3.14E-02 | 0.875514464 | 0.910565657 | 0.03444602 | 0.9725215 | NA | NA | ||||||||||||
| NA | NA | ||||||||||||||||||
| 95 conf int | NA | 95 conf int | NA | 95 conf int | |||||||||||||||
| Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | NA | Estimate | st SE | Adjusted SE | z value | Pr(>|z|) | |||
| (Intercept) | 39.34503249 | 1.92565228 | 1.99050365 | 19.76637044 | 0 | NA | (Intercept) | 6.700287577 | 0.44563973 | 0.46035289 | 14.55467701 | 0 | NA | (Intercept) | 26.9140026 | 2.16190181 | 2.23237161 | 12.05623762 | 0 |
| Axis.1 | -54.06305882 | 23.2473386 | 23.81149971 | 2.27046005 | 0.0231797 | NA | Axis.2 | -2.082292553 | 2.57543166 | 2.62119065 | 0.79440713 | 0.4269585 | NA | bio7 | 0.34177799 | 0.05653227 | 0.05838979 | 5.85338647 | 0 |
| Axis.2 | 105.8730629 | 88.45648472 | 89.42203144 | 1.18397067 | 0.2364247 | NA | bio6 | 0.430727698 | 0.03052731 | 0.03143785 | 13.70092854 | 0 | NA | Axis.1 | 1.57794905 | 11.89650289 | 12.18647211 | 0.12948366 | 0.896975 |
| Axis.3 | -5.432382412 | 26.52153131 | 27.54590024 | 0.19721201 | 0.8436616 | NA | Axis.1 | 0.22267537 | 1.25918711 | 1.2934862 | 0.17215133 | 0.8633186 | NA | Rearing.temp | 0.01174595 | 0.03343028 | 0.03422627 | 0.34318537 | 0.731459 |
| bio5 | 0.098505527 | 0.066665352 | 0.068957648 | 1.42849314 | 0.15315 | NA | Axis.4 | 0.171362227 | 2.27629114 | 2.35191008 | 0.07286088 | 0.9419168 | NA | Axis.2 | 4.24708937 | 26.46804905 | 27.17670287 | 0.15627684 | 0.8758148 |
| Axis.1:bio5 | 2.167761235 | 0.774452089 | 0.79221653 | 2.73632417 | 0.006213 | NA | Rearing.temp | -0.001971928 | 0.01542466 | 0.01593332 | 0.12376128 | 0.9015043 | NA | Axis.3 | 2.55023555 | 39.50379104 | 40.44935619 | 0.06304762 | 0.9497286 |
| Axis.2:bio5 | -3.410786773 | 2.820916563 | 2.850966 | 1.19636179 | 0.2315554 | NA | Axis.3 | 0.344867152 | 5.82039656 | 5.9665753 | 0.05779985 | 0.9539081 | NA | Axis.4 | -43.46029486 | 204.0829434 | 207.3599239 | 0.20958869 | 0.8339887 |
| Rearing.temp | 0.001015674 | 0.008166926 | 0.008450716 | 0.12018797 | 0.9043342 | NA | Axis.2:bio6 | -0.028382406 | 0.36667178 | 0.37805671 | 0.07507447 | 0.9401555 | NA | Axis.4:bio7 | 1.32978885 | 6.25950379 | 6.36007272 | 0.20908391 | 0.8343827 |
| Axis.4 | 2.178787485 | 41.79217286 | 43.10770462 | 0.05054288 | 0.9596898 | NA | Axis.1:bio6 | -0.054579821 | 0.21631607 | 0.21965669 | 0.24847785 | 0.8037647 | NA | Axis.2:bio7 | -0.10008424 | 0.7756989 | 0.79685177 | 0.12559957 | 0.9000489 |
| Axis.3:bio5 | 0.037161754 | 0.968321351 | 1.006521472 | 0.03692097 | 0.970548 | NA | Axis.3:bio6 | 0.016466532 | 0.41755684 | 0.42796108 | 0.0384767 | 0.9693076 | NA | Axis.1:bio7 | -0.03381928 | 0.34694592 | 0.35551648 | 0.09512717 | 0.9242138 |
| Axis.4:bio5 | -0.06684784 | 1.259095332 | 1.298786073 | 0.05146948 | 0.9589514 | NA | Axis.4:bio6 | 0.070484176 | 1.61820244 | 1.66039021 | 0.04245037 | 0.9661397 | NA | Axis.3:bio7 | -0.08611658 | 0.98198428 | 1.00527754 | 0.08566448 | 0.9317331 |
Page 19: 2016-06-01 Variance partitioning: thermal tolerance breadth example
Partitioning variation into phylogenetic (Axes 1-4), ecological (Tmax or Tmin or TAR), and phylogenetic + ecological components using the varpart() function in the vegan R package:
- a+b= phylo
- b= shared
- c+b= ecological
- a = phylo independent of ecology
- c = ecology independent of phylo code for model construction:
#Ctmax#varparfull<-varpart(merg$Ctmax,~Axis.1+Axis.2+Axis.3+Axis.4,~bio5,data=merg)fulloutput
Partition table: Df R.squared Adj.R.squared Testable[a+b] = X1 4 0.90796 0.89713 TRUE[b+c] = X2 1 0.75637 0.74979 TRUE[a+b+c] = X1+X2 5 0.90814 0.89422 TRUEIndividual fractions [a] = X1|X2 4 0.14444 TRUE[b] 0 0.75270 FALSE[c] = X2|X1 1 -0.00291 TRUE[d] = Residuals 0.10578 FALSE---Use function 'rda' to test significance of fractions of interestLooking at plots
plot(full)
#global model: a+b+canova(rda(merg$Ctmax~Axis.1+Axis.2+Axis.3+Axis.4+bio5,data=merg))#fraction a+bab<-rda(merg$Ctmax~Axis.1+Axis.2+Axis.3+Axis.4,data=merg)anova(ab)#frac b+cbc<-rda(merg$Ctmax~bio5,data=merg)anova(bc)#fraction a (phylo)a<-rda(merg$Ctmax~Axis.1+Axis.2+Axis.3+Axis.4+Condition(bio5),data=merg)anova(a)#fraction c (eco)c<-rda(merg$Ctmax~Condition(Axis.1+Axis.2+Axis.3+Axis.4)+bio5,data=merg)anova(c)Only showing code for CTmax I also applied variance paritioning for Ctmin and thermal tolerance breadth
Summary of results: Proportion of variance assigned to each component
Please scroll right to see the whole table, this table is wide
| Trait | Independent.Phylogeny | Independent.Ecology | Phylogeny | Ecology | Phylogeny.and.Ecology | Full | Residual |
|---|---|---|---|---|---|---|---|
| Ctmax | 0.14 | 0 | 0.90 | 0.75 | 0.75 | 0.90 | 0.10 |
| Ctmin | 0 | 0.31 | 0.64 | 0.92 | 0.60 | 0.91 | 0.09 |
| Tolerance Breadth | 0 | 0.45 | 0.17 | 0.57 | 0.11 | 0.53 | 0.47 |
Note-Bolded values represents significant variance component. The combined phylogeny and ecology variance component can not be tested for significance, only indirectly measured. The ecological component is represented by Tmax for Ctmax, Tmin for CTmin, and TAR for tolerance breadth.
Different way to represent proportion of variance explained by each component
CTmax

CTmin

Notes from climate cascade meeting, 2016-06-01
I have meetings with SHC and NJG every week, so I'll start logging our discussions here
We talked about the analysis from the thermal niche paper:
- NJG and SHC don't have strong feelings about model averaging.
- FOr the 4 panel field vs phytotron CTmax and Ctmin figure, keep separate lines for each species
- For thermal tolerance breadth, make 1 line
- Include variance partitioning analysis: Estimate amount of variance that go into phylogenetic components, ecological component, and their shared component.
- For CTmax , perform a Levine's test on the raw residuals from the regression line for picea (field vs phytotron).
- NJG: What does the literature say? Do people compare field vs common garden often? Do people assay thermal tolerance in the field alone?
Writing this up
SHC suggestion for results: Talk about field, then phyto, then present thermal tolerance breadth for phytotron.
For the phytroton gxp paper:
- Remake boxplot to include Axis 2
Page 21: 2016-06-02. Levine's test for raw residuals
We(SHC) suspect that the variance in field samples (for CTmax) is larger than the ones in the phytotron for A. picea.
 In this fig, just look at the blue line. Left is field, right panel is phyto.
In this fig, just look at the blue line. Left is field, right panel is phyto.
There is a cline in the field samples, but the cline goes away when comparing similar Tmax range as phyto:
This is a re-analysis for the field samples

TO test differences in variances, we'll performa Levine's test on the raw residuals.
About the test: some background
**Using the car package in R
Raw residuals in long format
| Raw Residuals | Field_V_phyto |
|---|---|
| 1.2426873 | field |
| 1.0498107 | field |
| 0.1956326 | field |
| -0.8463195 | field |
| 1.4852558 | field |
| -0.2966277 | field |
| 0.7426873 | field |
| 0.8078586 | field |
| 0.6459408 | field |
| -0.8070045 | field |
| -0.2070045 | field |
| -0.6070045 | field |
| -0.2070045 | field |
| -0.1799154 | field |
| -1.1114907 | field |
| -0.8269702 | field |
| -1.0805318 | field |
| -0.1320043 | phyto |
| -0.4520043 | phyto |
| 0.0980829 | phyto |
| 0.0980829 | phyto |
| -0.1842485 | phyto |
| 0.0157515 | phyto |
| -0.2211002 | phyto |
| -0.3411002 | phyto |
| -0.2292049 | phyto |
| -0.3492049 | phyto |
| -0.3492049 | phyto |
| -0.1640741 | phyto |
| -0.1040741 | phyto |
| -0.2040741 | phyto |
| -0.0640741 | phyto |
| -0.0720043 | phyto |
| 0.1879957 | phyto |
| 0.4388998 | phyto |
| 0.0388998 | phyto |
| 0.2988998 | phyto |
| 0.2388998 | phyto |
| 0.2739434 | phyto |
| 0.4939434 | phyto |
| 0.5959259 | phyto |
| 0.5559259 | phyto |
| 0.3679957 | phyto |
| 0.6329521 | phyto |
| 0.0070044 | phyto |
| -0.5389433 | phyto |
| -0.5789433 | phyto |
| -0.3589433 | phyto |
Code:
library(car)#levene's testleveneTest(lt[,1],lt[,2])Levene's Test for Homogeneity of Variance (center = median) Df F value Pr(>F) group 1 16.299 0.0002028 *** 46 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1#visualizingboxplot(lt[,1]~lt[,2],ylab="raw residuals",las=1)
Summary: Yes, sig diff in variance between field and phyto.
Page 22: 2016-06-02. Brute force fitting nls() functions in R!!
I googled how to fit nls even when failing to converge in R and found this gem.
Basically, use nls2() to brute force fit curves. I have not tried it, but putting it here as a ref.
Page 23: 2016-06-02. Literature reference for thermal niche paper to help write manuscript
Probably not comprehensive, but here it is:
Thermal breadth = 1 if they analyze it, 0 if they don't.
Table:
| Type | Author | Year | Journal | Taxa | Rearing_acclimation.Temperature | Heat_tolerance_Trait | Cold_tolerance_trait | Thermal_Breadth | Locale |
|---|---|---|---|---|---|---|---|---|---|
| Meta-analysis | Addo-Bediako et al. | 2000 | Proceedings of the royal society b | Insects | NA | Global | |||
| Lab acclimation | Deere & Chown | 2006 | American Naturalist | Mites | 1; 5; 10; 15 | Locomotion | Locomotion | 1 | Southern Ocean |
| Field | Compton et al. | 2007 | Experimental marine biology and ecology | Bivalve | Ctmax | Ctmin | 1 | Europe | |
| Lab acclimation | Calosi et al. | 2008 | Biology letters | Beetles | 14.5;20.5 | Ctmax | Ctmin | 0 | Europe |
| Lab acclimation | Calosi et al. | 2008 | Journal of biogeography | Beetles | 14.5; 20.5 | Ctmax | Ctmin | 1 | Africa to Europe |
| Field | Sinervo et al. | 2010 | Science | Lizards | Tb | 0 | Mexico | ||
| Lab acclimation | Calosi et al. | 2010 | Journal of Animal Ecology | Beetles | 14.5; 20.5 | Ctmax | Ctmin | 1 | USA |
| Lab acclimation | Anert et al. | 2011 | Integrative and Comparative Biology | Plants | 20-24 | RGR | RGR | 1 | USA |
| Meta-analysis | Sunday et al. | 2011 | Proceedings of the royal society b | Terrestrial and Marine | Ctmax | Ctmin | 1 | Global | |
| Common garden | Overgaard et al. | 2011 | American Naturalist | Fruit Fly | 25;29 | Ctmax;KO | Ctmin;KO | 1 | Australia |
| Common garden | Krenek et al. | 2012 | Plosone | Paramecium | 22 | GR | GR | 1 | Europe |
| Meta-analysis | Grigg & Buckley | 2012 | Biology letters | Lizards | Ctmax | Ctmin | 1 | Global | |
| Short acclimation | Sheldon & Tewksbury | 2014 | Ecology | Beetles | 20 | Ctmax | Ctmin | 1 | North and Central America |
| Common garden | Sheth & Angert | 2014 | Evolution | Plants | 20-25 | RGR | RGR | 1 | North America |
| Meta-analysis | Khaliq et al. | 2014 | Proceedings of the royal society b | Birds and Mammal | Ctmax | Ctmin | 1 | Global | |
| Short acclimation | Sheldon et al. | 2015 | Global Ecology and Biogeography | Lizards | 29 | Ctmax | Ctmin | 1 | Argentina |
| Lab acclimation | Bonino et al. | 2015 | Zoology | Lizards | 20-40 | Ctmax | Ctmin | 1 | Argentina |
| Velasco et al. | 2016 | Journal of biogeography | NA | Central America | |||||
| Meta-analysis | Lancaster | 2016 | Nature Climate Change | Insects | Ctmax | Ctmin | 1 | Global | |
| Lab acclimation | Gutierrez-Pesquera et al. | 2016 | Journal of biogeography | Frogs (tadpoles) | 20 | Ctmax | Ctmin | 1 | Global |
Page 24: 2016-06-03. Proteome stability project: Organizational entry
Today is Amanda's last day, so sad. She was working on the proteome stability project. Here I'll log all the organizational info that I'll need in the future:
- Where are the samples stored: Amanda and I both transferred the gel pieces, native and total protein boxes per colony to the -80C downstairs.

- For the TMT labeling, what order will they be labeled? See table below
- What else needs to be done? Wai and Bethany will resuspend our tryptic peptides, take some out and run some of the samples on LTQ to see if we have peptides. If we have peptides, then Wai will do the labeling for us! Wow!
Organization table: 3 pogo and 3 rudis colonies treated across 10 temperatures, that will be TMT labelled. LTQ run means that a subsample will be taken out to run on mass spec to check for peptides. ug of sample indicates how much protein we have.
| Species | Replicate | Colony | Temperature | Sample.. | Sample.Label | TMT.Label | LTQ...assignment | LTQ.Run | ug.of.Sample |
|---|---|---|---|---|---|---|---|---|---|
| P. barbatus | 1 | WWR45 | 30.1 | 2 | P45-2 | 126 | 1 | Yes | 7.28 |
| P. barbatus | 1 | WWR45 | 36.0 | 3 | P45-3 | 127N | 2 | Yes | 7.21 |
| P. barbatus | 1 | WWR45 | 41.2 | 4 | P45-4 | 127C | 3 | 6.23 | |
| P. barbatus | 1 | WWR45 | 43.9 | 5 | P45-5 | 128N | 4 | 5.95 | |
| P. barbatus | 1 | WWR45 | 46.3 | 6 | P45-6 | 128C | 5 | 5.41 | |
| P. barbatus | 1 | WWR45 | 48.2 | 7 | P45-7 | 129N | 6 | Yes | 4.59 |
| P. barbatus | 1 | WWR45 | 50.3 | 8 | P45-8 | 129C | 7 | 4.43 | |
| P. barbatus | 1 | WWR45 | 55.1 | 9 | P45-9 | 130N | 8 | 3.63 | |
| P. barbatus | 1 | WWR45 | 61.2 | 10 | P45-10 | 130C | 9 | Yes | 3.03 |
| P. barbatus | 1 | WWR45 | 65.2 | 11 | P45-11 | 131 | 10 | 3.08 | |
| A. rudis | 1 | Duke 1 | 30.1 | 2 | ARD1-2 | 126 | 11 | 6.69 | |
| A. rudis | 1 | Duke 1 | 36.0 | 3 | ARD1-3 | 127N | 12 | Yes | 6.00 |
| A. rudis | 1 | Duke 1 | 41.2 | 4 | ARD1-4 | 127C | 13 | 5.85 | |
| A. rudis | 1 | Duke 1 | 43.9 | 5 | ARD1-5 | 128N | 14 | Yes | 5.44 |
| A. rudis | 1 | Duke 1 | 46.3 | 6 | ARD1-6 | 128C | 15 | 4.86 | |
| A. rudis | 1 | Duke 1 | 48.2 | 7 | ARD1-7 | 129N | 16 | 4.50 | |
| A. rudis | 1 | Duke 1 | 50.3 | 8 | ARD1-8 | 129C | 17 | Yes | 3.79 |
| A. rudis | 1 | Duke 1 | 55.1 | 9 | ARD1-9 | 130N | 18 | Yes | 3.65 |
| A. rudis | 1 | Duke 1 | 61.2 | 10 | ARD1-10 | 130C | 19 | 3.13 | |
| A. rudis | 1 | Duke 1 | 65.2 | 11 | ARD1-11 | 131 | 20 | 2.66 | |
| P. barbatus | 2 | WWRQ53 | 30.1 | 2 | P53-2 | 126 | 21 | 6.54 | |
| P. barbatus | 2 | WWRQ53 | 36.0 | 3 | P53-3 | 127N | 22 | 6.21 | |
| P. barbatus | 2 | WWRQ53 | 41.2 | 4 | P53-4 | 127C | 23 | Yes | 5.82 |
| P. barbatus | 2 | WWRQ53 | 43.9 | 5 | P53-5 | 128N | 24 | 5.26 | |
| P. barbatus | 2 | WWRQ53 | 46.3 | 6 | P53-6 | 128C | 25 | 4.82 | |
| P. barbatus | 2 | WWRQ53 | 48.2 | 7 | P53-7 | 129N | 26 | 4.51 | |
| P. barbatus | 2 | WWRQ53 | 50.3 | 8 | P53-8 | 129C | 27 | Yes | 4.02 |
| P. barbatus | 2 | WWRQ53 | 55.1 | 9 | P53-9 | 130N | 28 | 3.64 | |
| P. barbatus | 2 | WWRQ53 | 61.2 | 10 | P53-10 | 130C | 29 | 2.81 | |
| P. barbatus | 2 | WWRQ53 | 65.2 | 11 | P53-11 | 131 | 30 | 2.79 | |
| A. rudis | 2 | Yates 2 | 30.1 | 2 | ARY2-2 | 126 | 31 | 6.10 | |
| A. rudis | 2 | Yates 2 | 36.0 | 3 | ARY2-3 | 127N | 32 | Yes | 6.52 |
| A. rudis | 2 | Yates 2 | 41.2 | 4 | ARY2-4 | 127C | 33 | 5.95 | |
| A. rudis | 2 | Yates 2 | 43.9 | 5 | ARY2-5 | 128N | 34 | 5.26 | |
| A. rudis | 2 | Yates 2 | 46.3 | 6 | ARY2-6 | 128C | 35 | 4.74 | |
| A. rudis | 2 | Yates 2 | 48.2 | 7 | ARY2-7 | 129N | 36 | Yes | 4.49 |
| A. rudis | 2 | Yates 2 | 50.3 | 8 | ARY2-8 | 129C | 37 | 4.30 | |
| A. rudis | 2 | Yates 2 | 55.1 | 9 | ARY2-9 | 130N | 38 | 3.57 | |
| A. rudis | 2 | Yates 2 | 61.2 | 10 | ARY2-10 | 130C | 39 | 3.01 | |
| A. rudis | 2 | Yates 2 | 65.2 | 11 | ARY2-11 | 131 | 40 | Yes | 2.83 |
| P. barbatus | 3 | WWRQ8 | 30.1 | 2 | P8-2 | 126 | 41 | 7.52 | |
| P. barbatus | 3 | WWRQ8 | 36.0 | 3 | P8-3 | 127N | 42 | 7.28 | |
| P. barbatus | 3 | WWRQ8 | 41.2 | 4 | P8-4 | 127C | 43 | 6.57 | |
| P. barbatus | 3 | WWRQ8 | 43.9 | 5 | P8-5 | 128N | 44 | 5.79 | |
| P. barbatus | 3 | WWRQ8 | 46.3 | 6 | P8-6 | 128C | 45 | Yes | 5.33 |
| P. barbatus | 3 | WWRQ8 | 48.2 | 7 | P8-7 | 129N | 46 | Yes | 4.87 |
| P. barbatus | 3 | WWRQ8 | 50.3 | 8 | P8-8 | 129C | 47 | Yes | 4.52 |
| P. barbatus | 3 | WWRQ8 | 55.1 | 9 | P8-9 | 130N | 48 | 3.88 | |
| P. barbatus | 3 | WWRQ8 | 61.2 | 10 | P8-10 | 130C | 49 | 3.46 | |
| P. barbatus | 3 | WWRQ8 | 65.2 | 11 | P8-11 | 131 | 50 | Yes | 3.31 |
| A. rudis | 3 | Lex 13 | 30.1 | 2 | ARL13-2 | 126 | 51 | 6.26 | |
| A. rudis | 3 | Lex 13 | 36.0 | 3 | ARL13-3 | 127N | 52 | 5.90 | |
| A. rudis | 3 | Lex 13 | 41.2 | 4 | ARL13-4 | 127C | 53 | Yes | 5.58 |
| A. rudis | 3 | Lex 13 | 43.9 | 5 | ARL13-5 | 128N | 54 | 5.01 | |
| A. rudis | 3 | Lex 13 | 46.3 | 6 | ARL13-6 | 128C | 55 | 4.51 | |
| A. rudis | 3 | Lex 13 | 48.2 | 7 | ARL13-7 | 129N | 56 | Yes | 3.95 |
| A. rudis | 3 | Lex 13 | 50.3 | 8 | ARL13-8 | 129C | 57 | Yes | 3.83 |
| A. rudis | 3 | Lex 13 | 55.1 | 9 | ARL13-9 | 130N | 58 | 3.41 | |
| A. rudis | 3 | Lex 13 | 61.2 | 10 | ARL13-10 | 130C | 59 | 2.62 | |
| A. rudis | 3 | Lex 13 | 65.2 | 11 | ARL13-11 | 131 | 60 | 1.79 |
A note: showing actual temperature treatments from thermal cycler
| Thermocylcer.Actual.Temp | Temperature |
|---|---|
| 25.0 | 25 |
| 30.1 | 30 |
| 36.0 | 35 |
| 41.2 | 40 |
| 43.9 | 43 |
| 46.3 | 45 |
| 48.2 | 48 |
| 50.3 | 50 |
| 55.1 | 55 |
| 61.2 | 60 |
| 65.2 | 65 |
| 70.1 | 70 |
Page 25: 2016-06-03. ggplot reference, updating a figure from Page 20: 2016-06-02
For JSG gxp ms that SHC is writing. Adding axis 2 into boxplot for hsp40 basal xp.
code for manipulating data so that I convert different axes into factors! There is probably a better way of doing this, but...
mergy<-subset(merg,merg$Axis.2> -0.1) # excluding axis 2 samplessub<-subset(merg,merg$Axis.2< -0.1)# taking out samples separating axis 2sub$axis3_desig<-rep("zAxis 2 A. picea",3) #naming factors based on axis2mergy$axis3_desig<-ifelse(mergy$Axis.3<= -0.044,"North",ifelse(mergy$Axis.3> 0.05,"South","A. picea")) # axis 3 designations!mergy<-rbind(mergy,sub) # combine them!mergy<-mergy[-54,] # 54th row has an NAggplot settings I like:
T<-theme_bw()+theme(text=element_text(size=30),axis.text=element_text(size=30),legend.text=element_text(size=28),panel.grid.major=element_blank(),legend.position="none",panel.grid.minor.x = element_blank(),panel.grid = element_blank(),legend.key = element_blank())Code to make fig
meds <- c(by(mergy$B_40, mergy$axis3_desig, median))Axis3_b40_v3<-ggplot(data=mergy,aes(x=factor(axis3_desig),y=B_40,fill=factor(axis3_desig)))+geom_boxplot()+T+ ylab(expression(paste("Hsp40 basal expression (",2^paste(Delta,Delta,"CT"),")")))+ scale_x_discrete(expression(paste(italic(" A. rudis")," clade")),labels=expression(paste(italic(" A. picea ")," North "," South "," Axis 2")))+ scale_y_continuous(limits=c(-1,11),breaks=seq(0,11,1))+ scale_fill_manual(name = "", values = c("gray","deepskyblue4", "firebrick","purple"))+guides(fill=FALSE)Axis3_b40_v3highlighting part of code where I can incorporate math symbols into the y axis:
ylab(expression(paste("Hsp40 basal expression (",2^paste(Delta,Delta,"CT"),")")))Final fig
I did play around with the fig in ppt first

Page 26: 2016-06-03 What is a cell type?
I was having lunch with Federico and thought: When I go to seminars and cell biologists use markers to indicate cell types, how do they know? What exactly is a cell type?
I've seen scientists using 1 marker to say, this is a this type of cell.
And in our graduate seminar series, there is a group that studies the physiology of taste receptors. They could not ID specific cell types and that part of the biology was unknown (not the typ 1/2/3's, but the VNO? ).
So I thought: Why not try to do single cell transcriptomics for 10 cells per group (what you think is a group). Then I'd explicitly test for differences using a discriminant analysis, or classification analysis. This approach could lead to a more quantitative justification for designating cell types.
Then we imagined that a totipotent cell that differentiates into diverse cell types can also follow or resemble a phylogenetic tree.
Feder suggests to read:
Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis Gene expression divergence recapitulates the developmental hourglass model
Page 27: 2016-06-03. qPCR plate layout and using the loaner ABI steponeplus Page 11: 2016-05-18
I started up the aBI steponeplus loaner today.

My usual 96 well plate layout is in my physical notebook, but I'll share it here:
| X | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Colony1:T1 | Colony1:T2 | Colony1:T3 | Colony1:T4 | Colony1:T5 | Colony1:T6 | Colony1:T7 | Colony1:T8 | Colony1:T9 | Colony1:T10 | Colony1:T11 | Colony1:T12 |
| B | Colony1:T1 | Colony1:T2 | Colony1:T3 | Colony1:T4 | Colony1:T5 | Colony1:T6 | Colony1:T7 | Colony1:T8 | Colony1:T9 | Colony1:T10 | Colony1:T11 | Colony1:T12 |
| C | Colony2:T1 | Colony2:T2 | Colony2:T3 | Colony2:T4 | Colony2:T5 | Colony2:T6 | Colony2:T7 | Colony2:T8 | Colony2:T9 | Colony2:T10 | Colony2:T11 | Colony2:T12 |
| D | Colony2:T1 | Colony2:T2 | Colony2:T3 | Colony2:T4 | Colony2:T5 | Colony2:T6 | Colony2:T7 | Colony2:T8 | Colony2:T9 | Colony2:T10 | Colony2:T11 | Colony2:T12 |
| E | Colony3:T1 | Colony3:T2 | Colony3:T3 | Colony3:T4 | Colony3:T5 | Colony3:T6 | Colony3:T7 | Colony3:T8 | Colony3:T9 | Colony3:T10 | Colony3:T11 | Colony3:T12 |
| F | Colony3:T1 | Colony3:T2 | Colony3:T3 | Colony3:T4 | Colony3:T5 | Colony3:T6 | Colony3:T7 | Colony3:T8 | Colony3:T9 | Colony3:T10 | Colony3:T11 | Colony3:T12 |
| G | Colony4:T1 | Colony4:T2 | Colony4:T3 | Colony4:T4 | Colony4:T5 | Colony4:T6 | Colony4:T7 | Colony4:T8 | Colony4:T9 | Colony4:T10 | Colony4:T11 | Colony4:T12 |
| H | Colony4:T1 | Colony4:T2 | Colony4:T3 | Colony4:T4 | Colony4:T5 | Colony4:T6 | Colony4:T7 | Colony4:T8 | Colony4:T9 | Colony4:T10 | Colony4:T11 | Colony4:T12 |
For each plate, I can run the 12 points of a performance curves for 4 colonies in duplicates. Each colony takes up 24 wells: 12 (T1-T12) temperature treatments and then ran in duplicates. Conditions for qpcr found here.
Usual temperatures:
- T1 - 25 C
- T2 - 28 C
- T3 - 30 C
- T4 - 31.5 C
- T5 - 33 C
- T6 - 35 C
- T7 - 36.5 C
- T8 - 38.5 C
- T9 - 40 C
- T10 - 41 C
- T11 - 25 C (middle of run)
- T12 - 25 C (end of run)
T11 and T12 are in there to serve as a time control (When I do the delta delta CT calculation, I'll include those to wash out the effect of time.
Actual samples I ran today for hsp40 541-641 primer pair, 55 C annealing.
Colonies:
- Duke2
- HF2
- Kite4
- Kite8
Summary of results:
All colonies had double peaks. So they're not usable. For these colonies, only hsp83 279-392 prims worked. Next, do 18s rRNA for housekeeping gene.
Silvia asked me to show her how to isolate RNA next Monday (2016-06-03), so I can isoalte CJ8( a colony I thought I isolated RNA from, but I didn't). It is in box 54
Page 28: 2016-06-03. Papers showing differences between fast static vs slow dynamic temperature treatments.
There is a large argument in the literature about how to best temperature treat ectotherms. One thing to point out, many fruit fly studies plot their heat tolerance traits against latitude, why not against local temperatures (Tmax, MAT)?
Here are a list of papers that find no clinal variation for slow ramp, but do for fast static experiments.
Sgro et al. 2010; Journal of Evolutionary Biology shows complex patterns between slow, hardening, and fast heat shocks across latitude.
Our group has argued that different temperature treatments represent different aspects of their thermal biology. Meaning:
- Fast heat shocks, whether dynamic or static = basal heat tolerance
- Slow heat shocks, whether dynamic or static = phenotypic plastic response in heat tolerance or acclimation or partial hardening response.
Page 29: 2016-06-06. Isolating RNA: colony CJ8; showing Sylvia
Isolated RNA and converted to cDNA. link to protocols
Colony: CJ8
Samples:
- CJ8 25
- CJ8 28
- CJ8 30
- CJ8 31.5
- CJ8 33
- CJ8 35
- CJ8 36.5
- CJ8 38.5
- CJ8 40
- CJ8 41
- CJ8 mid
- CJ8 last
- CJ8 25 -2
- CJ8 41 -2
Results of RNA isolation: we have RNA, now convert 50ng to cDNA
| N | Date | species | colony | box.condition | temp | Qubit_quant | conversion | Water.to.add |
|---|---|---|---|---|---|---|---|---|
| 799 | 20160606 | fulva | CJ8 | box54 | 25 | 11.70 | 4.27 | 5.73 |
| 800 | 20160606 | fulva | CJ8 | box54 | 28 | 11.50 | 4.35 | 5.65 |
| 801 | 20160606 | fulva | CJ8 | box54 | 30 | 6.19 | 8.08 | 1.92 |
| 802 | 20160606 | fulva | CJ8 | box54 | 31.5 | 30.50 | 1.64 | 8.36 |
| 803 | 20160606 | fulva | CJ8 | box54 | 33 | 41.40 | 1.21 | 8.79 |
| 804 | 20160606 | fulva | CJ8 | box54 | 35 | 43.30 | 1.15 | 8.85 |
| 805 | 20160606 | fulva | CJ8 | box54 | 36.5 | 19.70 | 2.54 | 7.46 |
| 806 | 20160606 | fulva | CJ8 | box54 | 38.5 | 14.20 | 3.52 | 6.48 |
| 807 | 20160606 | fulva | CJ8 | box54 | 40 | 34.00 | 1.47 | 8.53 |
| 808 | 20160606 | fulva | CJ8 | box54 | 41 | 12.20 | 4.10 | 5.90 |
| 809 | 20160606 | fulva | CJ8 | box54 | mid | 46.20 | 1.08 | 8.92 |
| 810 | 20160606 | fulva | CJ8 | box54 | last | 16.90 | 2.96 | 7.04 |
| 811 | 20160606 | fulva | CJ8 | box54 | 25_2 | 20.20 | 2.48 | 7.52 |
| 812 | 20160606 | fulva | CJ8 | box54 | 41_2 | 27.60 | 1.81 | 8.19 |
Master mix for cDNA conversion
| cDNA.synthesis | X1.rxn | X17.rxns |
|---|---|---|
| 10xBuffer | 2.0 | 34.0 |
| dNTP | 0.8 | 13.6 |
| multiscribe RT | 1.0 | 17.0 |
| Rnase | 1.0 | 17.0 |
| Primer | 2.0 | 34.0 |
| H20 | 3.2 | 54.4 |
| total rxn | 10.0 | 170.0 |
Steps:
- Put pcr strip tubes on ice.
- Add h20 specified above table
- Add RNA specified above table. Final volume should be 10uL
- Aliquot 10 uL of master mix to all tubes.
- PCR, see protocol link at beginning of post.
Page 30: 2016-06-07. Brute force fitting nls function in R revisited Page 22: 2016-06-02
I tried this on my desktop to play with the data quick and dirty, but it should go in my dissertation repo:
So the main problem I had in the past was that nls would stop if it the fit was poor, nls2() will brute force fit curves.
Here is my mock dataset:
knitr::kable(m)Colony | temp| FC_hsc701468| FC_Hsp83279| FC_Hsp831583| FC_hsp40424| T| |:------|----:|-------------:|------------:|-------------:|------------:|----:| |SHC6 | 25.0| 0.8180765| 1.2727190| 1.3741141| 1.5064240| 25.0| |SHC6 | 28.0| 0.8999074| 1.3778736| 2.3077710| 1.9297926| 28.0| |SHC6 | 30.0| 0.7922560| 0.9294879| 1.1390051| 1.2217515| 30.0| |SHC6 | 31.5| 0.8561583| 1.1546421| 0.8679142| 1.0613295| 31.5| |SHC6 | 33.0| 3.3855425| 1.9787656| 1.8540116| 2.6265211| 33.0| |SHC6 | 35.0| 7.1917199| 2.5450325| 3.5009441| 4.0735230| 35.0| |SHC6 | 36.5| 19.4708137| 3.4314556| 4.3630936| 5.6521932| 36.5| |SHC6 | 38.5| 30.8610304| 4.2174121| 6.7580144| 6.2792960| 38.5| |SHC6 | 40.0| 32.5603639| 4.6504188| 7.5401674| 9.2319657| 40.0| |SHC6 | 41.0| 26.0984907| 2.8898872| NA| 7.1626251| 41.0| |Avon | 25.0| 1.1732547| 1.2784472| 1.1390452| 1.1012987| 25.0| |Avon | 28.0| 1.4387152| 1.5022087| 1.3336969| 1.3226495| 28.0| |Avon | 30.0| 0.8752047| 1.1583008| 1.2902416| 0.7690966| 30.0| |Avon | 31.5| 1.1998622| 1.0781117| 1.2132109| 0.7942291| 31.5| |Avon | 33.0| 2.0881946| 2.1492356| 2.6482339| 1.8422990| 33.0| |Avon | 35.0| 6.7926522| NA| 4.8219890| 3.9911963| 35.0| |Avon | 36.5| 10.7125651| 4.0352515| 5.5000395| 4.0940620| 36.5| |Avon | 38.5| 22.8261858| 7.0736972| 9.0038236| 6.7629965| 38.5| |Avon | 40.0| NA| NA| NA| NA| 40.0| |Avon | 41.0| 32.0884860| 10.1245880| 12.7812078| 11.7503167| 41.0| |KH7 | 25.0| 0.8116304| 0.7712080| 0.9304326| 0.8853779| 25.0| |KH7 | 28.0| 1.0896696| 1.1911849| 1.1219525| 0.9427790| 28.0| |KH7 | 30.0| 1.1757139| 1.2275952| 1.3403029| 0.9426073| 30.0| |KH7 | 31.5| 1.3429711| 2.1066143| 1.7442530| 1.1386698| 31.5| |KH7 | 33.0| 3.7095882| 3.2454970| 2.8123354| 1.9888572| 33.0| |KH7 | 35.0| 7.6945833| 3.1906332| 3.0515822| 2.2617349| 35.0| |KH7 | 36.5| 19.6792961| 7.5792950| 5.6249460| 4.7316773| 36.5| |KH7 | 38.5| 25.7475125| 6.0603869| 5.5386550| 4.9766214| 38.5| |KH7 | 40.0| 47.1850131| 12.1240032| 9.7991379| 8.5075648| 40.0| |KH7 | 41.0| 44.5367758| 11.4567417| 9.6551695| 9.7848318| 41.0| |test | 25.0| 1.0000000| 10.0000000| 5.0000000| 1.0000000| 25.0| |test | 28.0| 1.0000000| 9.0000000| 5.0000000| 2.0000000| 28.0| |test | 30.0| 2.0000000| 8.0000000| 5.0000000| 4.0000000| 30.0| |test | 31.5| 3.0000000| 7.0000000| 5.0000000| 6.0000000| 31.5| |test | 33.0| 4.0000000| 6.0000000| 5.0000000| 8.0000000| 33.0| |test | 35.0| 5.0000000| 5.0000000| 5.0000000| 8.0000000| 35.0| |test | 36.5| 6.0000000| 4.0000000| 5.0000000| 8.0000000| 36.5| |test | 38.5| 7.0000000| 3.0000000| 5.0000000| 8.0000000| 38.5| |test | 40.0| 8.0000000| 2.0000000| 5.0000000| 8.0000000| 40.0| |test | 41.0| 9.0000000| 1.0000000| 5.0000000| 8.0000000| 41.0|
- Now I have to fit curves (boltzmann function) to for each colony and each gene (FC70, FC83, and FC_40). You can see I have a test colony with made up numbers, these should be poor fits.
I'm using nls2() and this curve estimates the critical temperature (Tm), slope (a), and max expression
Boltz<-function(data=x){ B<-nls2(gxp ~ (1+(max-1)/(1+exp((Tm-T)/a))),data=data, start=list(max=80,Tm=35,a=1.05), trace=TRUE,control=nls.control(warnOnly = TRUE, tol = 1e-05, maxiter=1000))#summary(B) return(summary(B)$parameters)}- I'll need to convert it long format, it is in wide right now.
names(m)[1] "Colony" "temp" "FC_hsc70_1468" "FC_Hsp83_279" [5] "FC_Hsp83_1583" "FC_hsp40_424" "T" > mlong<-gather(m,gene,gxp,FC_hsc70_1468:FC_hsp40_424)- fit for each colony and gene with ddply + Boltz functions
fits<-ddply(mlong,.(Colony,gene),Boltz)fits<-cbind(fits,rep(c("max","Tm","slope"),length(fits$Colony))) # adding parameter columnnames(fits)[7]<-"parameter"# renaming columnknitr::kable(fits)Won'tfit with test colony
Trying fits by removing test colony
mlong<-subset(mlong,mlong$Colony!="test")fits<-ddply(mlong,.(Colony,gene),Boltz)Output table!
| Colony | gene | Estimate | Std. Error | t value | Pr(>|t|) | parameter |
|---|---|---|---|---|---|---|
| Avon | FC_hsc70_1468 | 35.8189402 | 1.3830780 | 25.897990 | 0.0000002 | max |
| Avon | FC_hsc70_1468 | 37.7704625 | 0.1824726 | 206.992555 | 0.0000000 | Tm |
| Avon | FC_hsc70_1468 | 1.5075619 | 0.1117296 | 13.492950 | 0.0000103 | slope |
| Avon | FC_Hsp83_279 | 13.0621490 | 1.7746986 | 7.360207 | 0.0007271 | max |
| Avon | FC_Hsp83_279 | 38.5802879 | 0.7637267 | 50.515830 | 0.0000001 | Tm |
| Avon | FC_Hsp83_279 | 2.1031077 | 0.3554831 | 5.916195 | 0.0019659 | slope |
| Avon | FC_Hsp83_1583 | 16.8751069 | 2.4307114 | 6.942456 | 0.0004429 | max |
| Avon | FC_Hsp83_1583 | 38.4508017 | 0.9001894 | 42.714125 | 0.0000000 | Tm |
| Avon | FC_Hsp83_1583 | 2.4352914 | 0.3611821 | 6.742558 | 0.0005186 | slope |
| Avon | FC_hsp40_424 | 21.9643380 | 12.1034762 | 1.814713 | 0.1194923 | max |
| Avon | FC_hsp40_424 | 40.8933831 | 2.9441107 | 13.889893 | 0.0000087 | Tm |
| Avon | FC_hsp40_424 | 2.6054162 | 0.6918408 | 3.765919 | 0.0093334 | slope |
| KH7 | FC_hsc70_1468 | 57.0478157 | 12.0292674 | 4.742418 | 0.0021020 | max |
| KH7 | FC_hsc70_1468 | 38.2671391 | 1.0235944 | 37.385060 | 0.0000000 | Tm |
| KH7 | FC_hsc70_1468 | 1.7874874 | 0.5009719 | 3.568039 | 0.0091208 | slope |
| KH7 | FC_Hsp83_279 | 18.8164697 | 14.2023236 | 1.324887 | 0.2268193 | max |
| KH7 | FC_Hsp83_279 | 39.5972751 | 5.0039209 | 7.913250 | 0.0000977 | Tm |
| KH7 | FC_Hsp83_279 | 2.9760831 | 1.4783205 | 2.013152 | 0.0839745 | slope |
| KH7 | FC_Hsp83_1583 | 16.7337144 | 10.3102857 | 1.623012 | 0.1486163 | max |
| KH7 | FC_Hsp83_1583 | 40.1004665 | 4.0390733 | 9.928135 | 0.0000224 | Tm |
| KH7 | FC_Hsp83_1583 | 3.0388325 | 1.0845309 | 2.801979 | 0.0264489 | slope |
| KH7 | FC_hsp40_424 | 19.9496194 | 14.9270787 | 1.336472 | 0.2231999 | max |
| KH7 | FC_hsp40_424 | 41.3533804 | 3.8900811 | 10.630467 | 0.0000143 | Tm |
| KH7 | FC_hsp40_424 | 2.6777066 | 0.8223794 | 3.256048 | 0.0139403 | slope |
| SHC6 | FC_hsc70_1468 | 30.1357724 | 1.3518947 | 22.291509 | 0.0000001 | max |
| SHC6 | FC_hsc70_1468 | 36.0181917 | 0.2145002 | 167.916817 | 0.0000000 | Tm |
| SHC6 | FC_hsc70_1468 | 0.7601739 | 0.1966529 | 3.865562 | 0.0061670 | slope |
| SHC6 | FC_Hsp83_279 | 3.9378751 | 0.3837209 | 10.262341 | 0.0000180 | max |
| SHC6 | FC_Hsp83_279 | 34.4580183 | 0.8580317 | 40.159376 | 0.0000000 | Tm |
| SHC6 | FC_Hsp83_279 | 1.2755059 | 0.6850160 | 1.862009 | 0.1049016 | slope |
| SHC6 | FC_Hsp83_1583 | 8.6530046 | 1.6923497 | 5.113012 | 0.0021932 | max |
| SHC6 | FC_Hsp83_1583 | 36.6782852 | 1.1214736 | 32.705437 | 0.0000001 | Tm |
| SHC6 | FC_Hsp83_1583 | 1.8095631 | 0.6243422 | 2.898352 | 0.0273933 | slope |
| SHC6 | FC_hsp40_424 | 8.3707957 | 1.0694746 | 7.827017 | 0.0001048 | max |
| SHC6 | FC_hsp40_424 | 35.6669753 | 0.9166608 | 38.909679 | 0.0000000 | Tm |
| SHC6 | FC_hsp40_424 | 1.8169999 | 0.6708063 | 2.708680 | 0.0302567 | slope |
looks like it works when there is no poor fit.
Ok, I figured out how to suppress errors and let the funciton loop with failwith() function.
m<-read.csv("20160607_gxp_test.csv")m$T<-m$tempstr(m)#change to long formatmlong<-gather(m,gene,gxp,FC_hsc70_1468:FC_hsp40_424)str(mlong)#mlong<-subset(mlong,mlong$Colony!="test")fits<-ddply(mlong,.(Colony,gene),failwith(f=Boltz)) ## the magical code hereTable of outputs
| Colony | gene | Estimate | Std. Error | t value | Pr(>|t|) |
|---|---|---|---|---|---|
| Avon | FC_hsc70_1468 | 35.8189402 | 1.3830779 | 25.897991 | 0.0000002 |
| Avon | FC_hsc70_1468 | 37.7704625 | 0.1824726 | 206.992559 | 0.0000000 |
| Avon | FC_hsc70_1468 | 1.5075619 | 0.1117296 | 13.492950 | 0.0000103 |
| Avon | FC_Hsp83_279 | 13.0621489 | 1.7746986 | 7.360207 | 0.0007271 |
| Avon | FC_Hsp83_279 | 38.5802879 | 0.7637267 | 50.515830 | 0.0000001 |
| Avon | FC_Hsp83_279 | 2.1031077 | 0.3554832 | 5.916195 | 0.0019659 |
| Avon | FC_Hsp83_1583 | 16.8751071 | 2.4307113 | 6.942456 | 0.0004429 |
| Avon | FC_Hsp83_1583 | 38.4508017 | 0.9001893 | 42.714127 | 0.0000000 |
| Avon | FC_Hsp83_1583 | 2.4352914 | 0.3611821 | 6.742558 | 0.0005186 |
| Avon | FC_hsp40_424 | 21.9649309 | 12.1044659 | 1.814614 | 0.1195088 |
| Avon | FC_hsp40_424 | 40.8935313 | 2.9442708 | 13.889188 | 0.0000087 |
| Avon | FC_hsp40_424 | 2.6054554 | 0.6918546 | 3.765900 | 0.0093336 |
| KH7 | FC_hsc70_1468 | 57.0473854 | 12.0288922 | 4.742530 | 0.0021017 |
| KH7 | FC_hsc70_1468 | 38.2671031 | 1.0235676 | 37.386005 | 0.0000000 |
| KH7 | FC_hsc70_1468 | 1.7874685 | 0.5009659 | 3.568045 | 0.0091207 |
| KH7 | FC_Hsp83_279 | 18.8160754 | 14.2013489 | 1.324950 | 0.2267995 |
| KH7 | FC_Hsp83_279 | 39.5971341 | 5.0036704 | 7.913618 | 0.0000977 |
| KH7 | FC_Hsp83_279 | 2.9760359 | 1.4782900 | 2.013161 | 0.0839733 |
| KH7 | FC_Hsp83_1583 | 16.7333374 | 10.3095588 | 1.623090 | 0.1485996 |
| KH7 | FC_Hsp83_1583 | 40.1003166 | 4.0388773 | 9.928580 | 0.0000224 |
| KH7 | FC_Hsp83_1583 | 3.0387896 | 1.0845105 | 2.801992 | 0.0264484 |
| KH7 | FC_hsp40_424 | 19.9504446 | 14.9288152 | 1.336372 | 0.2232310 |
| KH7 | FC_hsp40_424 | 41.3536013 | 3.8903675 | 10.629742 | 0.0000143 |
| KH7 | FC_hsp40_424 | 2.6777587 | 0.8223999 | 3.256030 | 0.0139406 |
| Phil | FC_hsc70_1468 | 14.4816051 | 0.6238735 | 23.212404 | 0.0000028 |
| Phil | FC_hsc70_1468 | 34.8148669 | 0.2209902 | 157.540295 | 0.0000000 |
| Phil | FC_hsc70_1468 | 0.8480438 | 0.2387966 | 3.551322 | 0.0163645 |
| Phil | FC_Hsp83_279 | 4.6238796 | 0.4489827 | 10.298570 | 0.0001484 |
| Phil | FC_Hsp83_279 | 33.7411733 | 0.7422000 | 45.461025 | 0.0000001 |
| Phil | FC_Hsp83_279 | 1.2133128 | 0.5981040 | 2.028598 | 0.0982866 |
| Phil | FC_hsp40_424 | 4.3629872 | 0.2614315 | 16.688838 | 0.0000141 |
| Phil | FC_hsp40_424 | 34.6387089 | 0.3401929 | 101.820776 | 0.0000000 |
| Phil | FC_hsp40_424 | 0.7043699 | 0.3427897 | 2.054816 | 0.0950582 |
| SHC6 | FC_hsc70_1468 | 30.1357991 | 1.3519005 | 22.291433 | 0.0000001 |
| SHC6 | FC_hsc70_1468 | 36.0181969 | 0.2145014 | 167.915909 | 0.0000000 |
| SHC6 | FC_hsc70_1468 | 0.7601800 | 0.1966547 | 3.865558 | 0.0061670 |
| SHC6 | FC_Hsp83_279 | 3.9379010 | 0.3837369 | 10.261982 | 0.0000180 |
| SHC6 | FC_Hsp83_279 | 34.4580679 | 0.8580653 | 40.157863 | 0.0000000 |
| SHC6 | FC_Hsp83_279 | 1.2755764 | 0.6850461 | 1.862030 | 0.1048984 |
| SHC6 | FC_Hsp83_1583 | 8.6530046 | 1.6923498 | 5.113012 | 0.0021932 |
| SHC6 | FC_Hsp83_1583 | 36.6782851 | 1.1214737 | 32.705435 | 0.0000001 |
| SHC6 | FC_Hsp83_1583 | 1.8095631 | 0.6243422 | 2.898351 | 0.0273933 |
| SHC6 | FC_hsp40_424 | 8.3707958 | 1.0694747 | 7.827016 | 0.0001048 |
| SHC6 | FC_hsp40_424 | 35.6669753 | 0.9166608 | 38.909677 | 0.0000000 |
| SHC6 | FC_hsp40_424 | 1.8169999 | 0.6708063 | 2.708680 | 0.0302567 |
| test | FC_hsc70_1468 | 9.8719349 | 0.9800918 | 10.072460 | 0.0000204 |
| test | FC_hsc70_1468 | 35.6649510 | 0.8966939 | 39.773830 | 0.0000000 |
| test | FC_hsc70_1468 | 2.9884380 | 0.4909301 | 6.087299 | 0.0004973 |
| test | FC_hsp40_424 | 8.0828867 | 0.1090835 | 74.098170 | 0.0000000 |
| test | FC_hsp40_424 | 30.3192228 | 0.1219349 | 248.650901 | 0.0000000 |
| test | FC_hsp40_424 | 1.1145318 | 0.1136478 | 9.806893 | 0.0000243 |
Notice:
That not all genes have fitted parameters! nice! ie. test hsp83's!
Now we need to:
- Predict new sets of values for each gene/colony
- Visualize actual vs predicted values!
Code to predict new values
- first, the plotting function
fud<-function(T=seq(25,70,.1),Tm=40,slope=1.8,max=50){ y<-1+ (max-1)/(1+exp(((Tm-T)/slope))) return(y) }plot(fud())- OK, now the data manipulation
#grab fitted lines from estimates#change to wide formatlibrary(reshape2)feeder<-dcast(fits2,Colony+gene~parameter,value.var="Estimate")list_predictions<-sapply(split(feeder,list(feeder$Colony,feeder$gene)),function(x) {fud(T=seq(25,45,.1),Tm=x$Tm,slope=x$slope,max=x$max)})predi<-as.data.frame(do.call("rbind", list_predictions),stringAsFactors=FALSE)predi$Sample<-row.names(predi)nom<-as.data.frame(matrix(unlist(strsplit(predi$Sample,"[.]")),ncol=2,byrow=TRUE)) #messing with the namesnames(nom)<-c("Colony","gene")predictions<-cbind(predi,nom)##gotta change to long formatconv<-gather(predictions,Colony,gxp,V1:V201)[,-4]#need to sortconv<-conv[order(conv$Sample),] #dont forget to order!!!plong<-cbind(conv,rep(seq(25,45,.1),nrow(predi)))names(plong)[5]<-"T"head(plong)Plotting with ggplot
for hsc70-4 h2
- lines = predicted fit from function
- points = empirical
b<-subset(plong,plong$gene=="FC_hsc70_1468")qplot(x=T,y=gxp,data=subset(mlong,mlong$gene=="FC_hsc70_1468"),colour=Colony)+geom_point(size=5)+xlim(25,45)+geom_point(aes(y=gxp,x=T,colour=Colony),data=b)
- hsp83 279

- hsp40 541

Page 31: 2016-06-08. Redoing online notebook template
I updated my online notebook template.. I probably should have done this from the start. But there is a table of contents with 200 entries with automatic links to those entries.
Code for automatically generating table of contents:
* [Page 1: Date](#id-section1). Title * [Page 2: Date](#id-section2). Title
For table of contents, you want this syntax:
- I used R with a series of paste functions to get the right syntax
- Exported to csv and just pasted it into the markdown
#constructing table of contentsone<-rep("* [Page",200)two<-seq(1:200)three<-paste(one,two)four<-paste(three,":","]",sep="")five<-paste(four,"(#id-section",two,").",sep="")six<-data.frame(five)write.csv(six,"ffff.csv")Code for automatically generating entries with titles that correspond with table of contents
For this you want this syntax:
------ <div id='id-section1'/>
- R manipulations
b<-rep("------",200)c<-rep("<div id='id-section",200)d<-seq(1:200)e<-paste(d,"'/> ",sep="")m<-paste(c,e,sep="")mi<-rep("### Page",200)i2<-paste(i,rep(1:200))i3<-paste(i2,":",sep="") # can even add year herem1<-paste(b,m,i3,sep=" ")write.csv(m1,"testy.csv")- Export to csv
- You do need to get rid of header and first column manually, save and close (in excel)
- Open in textwrangler and you'll see that the line breaks appear. Then get rid of quotes.
Page 32: 2016-06-08. qPCRs, 18s rRNA for Duke2, HF2, Kite 4, Kite8, 60 C annealing
Ran qpcr plate (96 well) on loaner ABI steponeplus. Samples were already 1/10 diluted, and for 18s, I dilute 1/10 again to have a 1/100 dilution.
Colonies:
- Duke2
- HF2
- Kite4
- Kite8
- Made master mix: added 550 uL sybr green, 21 uL F+R primer, and 84 uL h20
- Dispensed 6 uL into plate
- Added 4 uL of cDNA (1/100 dilution) into plate
- qPCR, 60 C annealing
Summary:
Single peaks from melt curve analysis indicating single amplicon. The threshold was set to 0.5.
Updated summary of whole project so far:
| Progress | X18s | hsc70.414681592_degen | hsp83279392_degen | hsp8315831682_degen | hsp40424525degen |
|---|---|---|---|---|---|
| works | 59 | 51 | 57 | 41 | 51 |
| double peaks | 2 | 11 | 5 | 19 | 7 |
| total | 61 | 62 | 62 | 60 | 58 |
Dilutions of future samples
Dilute 1/10: 5 uL of sample + 45 uL of h20 in 12 strip pcr tubes.
Sample colonies:
- CJ2
- CJ5
- Duke1
- SHC8
Page 33: 2016-06-08. Climate cascade meeting.
SHC can't make it. KM going to process samples. ANBE + NJG meet
- Evolution poster: Go over figures and conclusions
- Update gxp curve fitting
-
NJG suggestions:
- For figure4, gray out points and put pretreatment temps beside each line.
- Figure 3, plot hardening ability vs basal cold tolerance.
-
Page 34: 2016-06-09; 2016-06-10. qPCRs: Duke1, CJ2, SHC8, CJ5
- hsc70-4 h2 1468, 60C annealing results: only Duke1 worked
- hsc70-4 h2 1468, 55C annealing results: none worked
- hsp83 279 prim, 55C annealing results: all worked
- hsp40 541 prim, 55C annealing results: all worked , although some replicates excluded due to non-specificity
- 18s rRNA, 60 C annealing results: Samples were diluted 1/10.
Page 35: 2016-06-10. ABI steponeplus machine fix and sending back instrument.
machine repaired
*Dear Andrew, The repair of your instrument on service reference notification 405638599 has been completed and is now on its way back to you. For your record the reference tracking number is 650686939762 I will be sending you a separate email with the decontamination forms and FedEx labels to return the loaner you received during the repair of yor instrument. Please send this loaner back in a timely fashion as we do have other customers in need of this loaner. Thank you, Leticia C. Instrument Services
Life Sciences Solutions*
Sending back loaner
Dear Andrew,
Attached you will find the necessary paperwork to ensure that the loaner unit is returned correctly and promptly.
- Your RMA is 14635-69
- Please review and complete the attached decontamination form and print out 2 copies.
- Please remember to place the instrument in the "Ship Prep" position prior to packing the instrument.
- Please DO NOT include your power cord with your instrument (remove from unit and keep it).
- Please DO NOT include any consumables (trays, tubes, etc.).
- Place a copy of the completed decontamination form INSIDE and OUTSIDE of the box.
- Print out the FedEx label, (link will arrive via separate email).
The return transaction cannot be processed until the completed decontamination form and the instrument are received. Thank you, Leticia C. Instrument Services
Life Sciences Solutions
2016-06-13 update
We received the repaired machine back.
Here is the decomtamination form for the loaner.
Page 36: 2016-06-10. Thoughts on Kingsolver & Woods 2016, AmNat. ref here
reference:
- Kingsolver JG, Woods HA. 2016. Beyond Thermal Performance Curves: Modeling Time-Dependent Effects of Thermal Stress on Ectotherm Growth Rates. The American Naturalist 187:283–294.
This paper models growth rate under heat stress over time. The authors use Hsp gene and protein expression as a measure of cost and ingestion rate as a trait that inputs energy into an animal.
Fig 1:

The physiology is more complicated than this. First, increasing Hsp gene expression is costly in itself, so there should be a separate cost term. While the actual Hsp protein expression is costly to invest into too, there is a cost for using them and also having unstable proteomes. Also, organisms can get rid of unstable proteins through degradation and haulting translation which would offsets the costs of Hsp (gene or protein) expression and using it. Basically, I'm saying the actual cost incurred come in the form of macromolecular damage (proteome stability) and the response to macromolecular damage (Hsp expression). Not sure if proteome stability cost needs to included
But here is a fig for proteome stability (prop non-denatured) as a function of Temperature:

The black line is 10 min incubations, the red line is 20 min. I fit a non-linear logistic curve to it link. This captures the incurring costs associated with temperature AND time without an acclimation response. It'd be interesting to develop a model from this....
2016-06-11. Follow up model

I've included potentially important physiological components. Macromolecular damage includes unstable proteins and damage to membranes. For simplicity, it can just represent unstable proteins. On second thought, it should be macromolecular stability, assuming there is an optimal stability of membranes and proteins for growth. So temperature directly affects macromolecular stability and given a certain amount of damage(instability), it elicits a physiological response ( transcription + translation) . Transcription includes all the transcripts that turn on and turn off. If the net effect is using more energy to turn on/off over higher temperatures, this incurs a cost. Same with translation, but there is also a cost of "using" the proteins. For example, Hsp mediated folding uses ATP. However, the combination of altering translation rates and using the proteins offsets the costs of macromolecular damage which directly affects growth.
Anyway, I'd call this the "thermostat" model.
- Craig EA, Gross CA. 1991. Is hsp70 the cellular thermometer? Trends in Biochemical Sciences 16:135–140.
2016-06-13. Predictions of thermostat model
- There is some temperature where the costs associated with macromolecular damage exceeds any type of physiological response (transcription, translation), resulting in inhibited growth.
- Under sublethal temperature stress, the negative long term outcome of inducing a physiological response may be tempered by increasing ingestion rate.
- Although a physiological response is costly, at sublethal levels, the combination of gene/protein expression(downregulating unstable proteins) and upreg of Hsps may have a net positive effect on proteome stability, which is related to growth.
Note: There is a cool paper by Hoekstra & Montooth that shows how Hsp70 expression covaries with metabolic rate.
- Hoekstra LA, Montooth KL. 2013. Inducing extra copies of the Hsp70 gene in Drosophila melanogaster increases energetic demand. BMC Evolutionary Biology 13:68.
Other thoughts:
- One cool thing about the model is that you can add transcriptome, proteome data as parameters into the model. How?
- Count the costs of each transcript (# of basepairs) and subtract response from baseline to get relative response. One could argue that overall, Hsp expression is not costly because other transcripts can be downregulated at the same time. I don't think anybody has tried to explore this in transcriptome datasets.
- In aquatic systems, oxygen limitation seems to be the mechanism for upper thermal limits. Is there a way to make one global model so that we can make predictions for any ectotherm?
Page 37: 2016-06-11. Quantifying natural selection in natural populations
I've been reading more Kingsolver (specifically Kingsolver et al. 2001;Kingsolver & Diamond 2011 which led me to think about quantifying modes of selection in nature. Basically all you need to do is regress traits against relative fitness (fitness of individual / mean fitness of population). The slope is the magnitude of direction selection. Also, if you want to detect disruptive or stabilizing, then you can add a quadratic term. It'd be interesting to apply this technique to assess the fitness consequences of climate change. So take a species and measure fitness and traits along a transect to pick up the warm-edge, core, and cool-edge populations.
Refs for me to read:
- Lande & Arnold 1983
- Arnold & Ward 1984 for measuring natural and sexual selection
- Mitchell-Olds & Shaw 1987 for pitfalls in these regressions analyses.
- Heisler & Damuth 1987 for multilevel selection and it also introduces contextual analyses.
Page 38: 2016-06-13. qPCR update for Duke1,CJ2,SHC8,CJ5. Randomizing samples treated at 25C(reference for basal expression) for qpcrs.
Running qpcr for Duke1/CJ2/SHC8/CJ5; hsc70-4 h2 50C annealing.
Randomizing procedure
- Load data set as csv in R
- Code for sampling randomly:
write.csv(sample(d$colonies),"ra.csv")- Changed csv so that I have rows and columns
- Here is the layout:
| Row | Column | Colony |
|---|---|---|
| A | 1 | Ala1 |
| A | 2 | KITE8 |
| A | 3 | Yates2 |
| A | 4 | FBRAGG3 |
| A | 5 | CJ4 |
| A | 6 | BK |
| A | 7 | HW7 |
| A | 8 | KH3 |
| A | 9 | DUKE9 |
| A | 10 | SHC8 |
| A | 11 | CJ2 |
| A | 12 | HF2 |
| B | 1 | shc7 |
| B | 2 | MA |
| B | 3 | PB07-23 |
| B | 4 | CJ8 |
| B | 5 | Lex9 |
| B | 6 | ApGxL10A |
| B | 7 | Phillips |
| B | 8 | hf3 |
| B | 9 | PB17-10 |
| B | 10 | CJ6 |
| B | 11 | Ala4 |
| B | 12 | CJ5 |
| C | 1 | PB17-14 |
| C | 2 | DUKE8 |
| C | 3 | KH1 |
| C | 4 | Greenfield |
| C | 5 | fbragg1 |
| C | 6 | Avon19.1 |
| C | 7 | CampNSP |
| C | 8 | KH6 |
| C | 9 | KH5 |
| C | 10 | DUKE2 |
| C | 11 | SHC9 |
| C | 12 | LPR2 |
| D | 1 | KITE4 |
| D | 2 | FBRAGG4 |
| D | 3 | KH7 |
| D | 4 | DUKE1 |
| D | 5 | PMBE |
| D | 6 | DUKE6 |
| D | 7 | CJ7 |
| D | 8 | fbragg5 |
| D | 9 | CJ1 |
| D | 10 | LPR4 |
| D | 11 | YATES3 |
| D | 12 | POP1 |
| E | 1 | kh2 |
| E | 2 | Bingham |
| E | 3 | SHC3 |
| E | 4 | ApGxL09A |
| E | 5 | Ted6 |
| E | 6 | DUKE7 |
| E | 7 | SHC6 |
| E | 8 | DUKE4 |
| E | 9 | DUKE5 |
| E | 10 | Ted4 |
| E | 11 | EXIT65 |
| E | 12 | sidewalk (formica) |
| F | 1 | POP2 |
| F | 2 | fbragg2 |
| F | 3 | SHC2 |
| F | 4 | LEX13 |
| F | 5 | SHC5 |
| F | 6 | cremat |
| F | 7 | SHC10 |
| F | 8 | pop3 |
| F | 9 | SR45 |
| F | 10 | AS4 |
I'll arrange these samples in rows of 12 in pcr strip tubes, dilute 1/10 and then I can multichannel the samples into a 96 well qpcr plate.
Page 39: 2016-06-13. Post doc project idea: Assessing current impacts of climate change in natural populations.
Alternate title: Quantifying the intensity of selection associated with climate change.
Question: Are populations experiencing selection associated with climate change out in nature?
Hypothesis: The magnitude and direction of selection acts on different parts of their range depending on their thermal environment.
Predictions:
- Individuals at the warm edge of their range experience positive directional selection for a thermal trait.
- Individuals at the core experience stabilizing selection for a thermal trait.
- Individuals at the cool edge experience negative directional selection for a thermal trait.
Approach: Measure phenotypic selection on physiological, behavioral traits across a cline for a given species. A good system to measure phenotypic selection are ants because alates are direct measurement of fitness. So the product of # of alates by their weights will give a meaurement of fitness. Then, regress different traits on relative fitness to obtain a selection gradient. I can detect disruptive and stabilizing selection by adding a quadratic term in the regression model. I don't want to automatically assign individuals to warm-edge, cool edge, core. I'd sample along a cline (10-20 sites?). Also, there may be differences in the phenology for alates to develop, so I'd probably need to sample 3-4 times a year?
Some key traits:
- Colony size ( # of workers, # of larvae, # of pupae, Colony biomass really)
- Thermal tolerance ( CTmax, Ctmin, KO-time, hardening ability)
- Morphology ( leg length, average worker weight)
Some things to think about:
- I read somewhere (find it) that what one really wants is the life time reproductive success (LRS). But this is almost impossible to measure. In this sense, it is more accurate to say I'm measuring episodic selection (Angiletta 2009)?
- Also, one should be comparing within a generation. There may be different age classes of colonies, but it may be reasonable to assume that if the colony has alates, then they belong to a similar age class.
- I'd need to do some pop gen to determine the population level structure so that I can empirically assign individuals to populations.
Another thought: Phenotypic selection seems like a good way to associate higher and lower phenotypic levels.
- For example, I have CTmax data and the underlying stress response measured. CTmax is a component of fitness, so if I regress the stress response onto the relative fitness of CTmax(CTmax of individual/ population CTmax mean) , then I can determine a selection gradient.
- I can also measure phenotypic selection for allele frequencies! (Dr. Goodnight's suggestion)
Page 40: 2016-06-14. qPCR's: Diluting samples for quantifying basal expression and repeats
Diluting samples for basal expression:
I diluted 1x cDNA samples 1:10, so I added 5 uL cDNA with 45 uL water. I added 25C-mid samples (because of technical mistake in diluting) for some colonies to replace 25C samples that were started at the beginning of heat shock.
- F10: Duke8 41 (switched with AS4)
- F11: SHC10 mid
- F12: AS4 25C
- G1: yates3 mid
- G2: shc2 mid
- G3: exit65 mid
- G4: greenfield mid
I also diluted the 1:10 cDNA samples again at 1:10 to run 18s rRNA. So I added 2 uL cDNA into 18 uL water.
All in all, it took ~ 3 hours from organization to completion.
Updated plate layout:
| Row | Column | Colony |
|---|---|---|
| A | 1 | Ala1 |
| A | 2 | KITE8 |
| A | 3 | Yates2 |
| A | 4 | FBRAGG3 |
| A | 5 | CJ4 |
| A | 6 | BK |
| A | 7 | HW7 |
| A | 8 | KH3 |
| A | 9 | DUKE9 |
| A | 10 | SHC8 |
| A | 11 | CJ2 |
| A | 12 | HF2 |
| B | 1 | shc7 |
| B | 2 | MA |
| B | 3 | PB07-23 |
| B | 4 | CJ8 |
| B | 5 | Lex9 |
| B | 6 | ApGxL10A |
| B | 7 | Phillips |
| B | 8 | hf3 |
| B | 9 | PB17-10 |
| B | 10 | CJ6 |
| B | 11 | Ala4 |
| B | 12 | CJ5 |
| C | 1 | PB17-14 |
| C | 2 | DUKE8 |
| C | 3 | KH1 |
| C | 4 | Greenfield |
| C | 5 | fbragg1 |
| C | 6 | Avon19.1 |
| C | 7 | CampNSP |
| C | 8 | KH6 |
| C | 9 | KH5 |
| C | 10 | DUKE2 |
| C | 11 | SHC9 |
| C | 12 | LPR2 |
| D | 1 | KITE4 |
| D | 2 | FBRAGG4 |
| D | 3 | KH7 |
| D | 4 | DUKE1 |
| D | 5 | PMBE |
| D | 6 | DUKE6 |
| D | 7 | CJ7 |
| D | 8 | fbragg5 |
| D | 9 | CJ1 |
| D | 10 | LPR4 |
| D | 11 | YATES3 |
| D | 12 | POP1 |
| E | 1 | kh2 |
| E | 2 | Bingham |
| E | 3 | SHC3 |
| E | 4 | ApGxL09A |
| E | 5 | Ted6 |
| E | 6 | DUKE7 |
| E | 7 | SHC6 |
| E | 8 | DUKE4 |
| E | 9 | DUKE5 |
| E | 10 | Ted4 |
| E | 11 | EXIT65 |
| E | 12 | sidewalk (formica) |
| F | 1 | POP2 |
| F | 2 | fbragg2 |
| F | 3 | SHC2 |
| F | 4 | LEX13 |
| F | 5 | SHC5 |
| F | 6 | cremat |
| F | 7 | SHC10 |
| F | 8 | pop3 |
| F | 9 | SR45 |
| F | 10 | Duke 8 41 |
| F | 11 | SHC10 mid |
| F | 12 | AS4 |
| G | 1 | yates3 mid |
| G | 2 | shc2 mid |
| G | 3 | exit65 mid |
| G | 4 | gf mid |
Repeats ran alongside CJ8
Ran hsp83 279 55 C annealing for following coloines:
- Fbragg1
- CJ1
- CJ8
- KH1; 1 row
- FB4; 1 row
results: Fb4 not work
Ran hsp40 541 prim 55C annealing for the same colonies as above.
results: CJ8 and KH1 worked
Ran 18s rRNA for following colonies:
- CJ1
- CJ8
- KH1
results: all worked
Update of samples:
| Status | X18s | hsc70.414681592_degen | hsp83279392_degen | hsp8315831682_degen | hsp40424525degen |
|---|---|---|---|---|---|
| works | 67 | 58 | 65 | 45 | 57 |
| double peaks | 0 | 9 | 2 | 20 | 10 |
| total | 67 | 67 | 67 | 65 | 67 |
Page 41: 2016-06-15. qPCRs to quantify basal expression. (Evolution of stress response project)
I probably should have mentioned this earlier, but since all the samples are on 1 plate, I'll be quantifying 4 genes in a replicated randomized block design.
So for each gene, run 2 plates. Samples on the plate were already previously randomized.
- Ran 18s rRNA plate 1, 55 C annealing temp.
- Ran hsc70-4 h2 1468 plate 1, 55 C annealing temp.
- Ran hsp83 279 plate 1, 55 C annealing temp.
- Ran hsp40 541 plate 1, 55 C annealing temp.
Page 42: 2016-06-15. Evolution talks I want to attend.
Not a comprehensive list, but a start.
| Day | Speaker | Room | Time | Title | Session |
|---|---|---|---|---|---|
| Monday, June 20 | Tangwancheroen, Sumaetee | MR10C | 1:30PM | Adaptation via divergence in gene regulation along a temperature cline: cis and trans effects on HSP expression the copepod Tigriopus californicus | Adaptation 1 |
| Monday, June 20 | Lyons,Marta | BallroomC | 2:00PM | Predicting range contractions in niche conserved plethodontid salamanders comparing correlative and biophysical niche models | Evolutionary ecology 1 |
| Saturday, June 18 | Gilbert, Kimberly | MR6B | 1:30PM | Local maladaptation interacts with expansion load during species range expansions | Population genetics theory methods 1 |
| Saturday, June 18 | Kingsolver,Joel | BallroomC | 9:15AM | Elevational clines in plastic and evolutionary responses of montane butterflies to climate change | Contemporary evolution |
| Sunday, June 19 | Nunney,Leonard | MR9AB | 2:45PM | Adapting to a changing environment: modeling the interaction of directional evolution and plasticity | Phenotypic plasticity |
| Sunday, June 19 | Muir,Chris | BallroomA | 8:30AM | What is evolutionary physiology? | Evolutionary physiological synthesis 1 |
| Sunday, June 19 | Garcia,Matteo | MR7 | 9:00AM | Performance determines division of labor in leafcutting ants | Social systems 1 |
| Sunday, June 19 | Campbell Staton, Shane | MR9C | 9:15AM | Polar Vortex cold wave elicits rapid physiological, regulatory and genetic shifts in populations of the green anole, Anolis carolinensis | Expression studies |
| Sunday, June 19 | Fumagalli, Sarah | MR7 | 9:30AM | The evolution of cooperation between unrelated individuals | Social systems 1 |
| Sunday, June 19 | Catullo,Renee | BallroomC | 10:15AM | Extending spatial modelling of climate change responses beyond the realized niche: estimating, and accommodating, physiological limits and adaptive evolution | Niche modeling |
| Sunday, June 19 | Powell,Scott | MR9AB | 10:15AM | Diversification of complex social phenotypes: insights from the turtle ants | Adaptation |
| Sunday, June 19 | Sexton, Jason | MR6A | 10:45AM | Does species niche breadth predict plant performance in novel environments? An experimental test in Australian Alps plants | Biogeography I |
| Sunday, June 19 | Rosauer,Dan | BallroomC | 10:45AM | Distribution models below species level | Niche modeling |
| Sunday, June 19 | Chau,Linh | MR7 | 10:45AM | Gene Duplication in the Evolution of Sex- and Caste-biased Gene Expression in Social Insects | Social systems 2 |
| Sunday, June 19 | Gunderson,Alex | BallroomA | 11:00AM | The physiology of adaptive radiation | Evolutionary physiological synthesis 2 |
| Sunday, June 19 | Angert,Amy | BallroomA | 11:15AM | Linking physiology to biogeography in monkeyflowers | Evolutionary physiological synthesis 2 |
| Sunday, June 19 | Parker,Joseph | MR9AB | 11:15AM | An inordinate fondness for rove beetles: evolution and diversification of ant social parasites | Adaptation |
Page 43: 2016-06-16. Figure for curve fitting: see Success with failwith() and Status update of samples.
Hsp70, 40, 83 from top to bottom

Page 44: 2016-07-18. Summary statistics for modulation of Hsp paper.
Overall means
| mean xp | |
|---|---|
| FC_83 | 11.218868 |
| FC_70 | 50.227915 |
| FC_40 | 10.535062 |
| B_83 | 1.735492 |
| B_70 | 1.446917 |
| B_40 | 1.935067 |
Comparison among genes

medians
| Rearing_Temp | Induction83 | Basal83 | Induction70 | Basal70 | Induction40 | Basal40 |
|---|---|---|---|---|---|---|
| 20 | 7.046216 | 0.9032384 | 48.88187 | 0.4773797 | 6.903618 | 1.155806 |
| 26 | 10.441149 | 1.5197949 | 39.13139 | 2.2318297 | 13.267033 | 1.559372 |
means
| Rearing_Temp | Induction83 | Basal83 | Induction70 | Basal70 | Induction40 | Basal40 |
|---|---|---|---|---|---|---|
| 20 | 9.352522 | 1.262254 | 55.45230 | 0.640272 | 8.059647 | 1.680941 |
| 26 | 14.320365 | 2.334319 | 42.62233 | 2.565149 | 14.086744 | 2.299683 |
Page 45: 2016-07-19. Meeting with VGN proteomics facility
Meeting with Wai and Bethany to finish up the comparative proteomics project (Amanda was working on this).
I went over our experimental protocol. Wai suggested to do searches with MASCOT and SEQUEST to ID more proteins.
Timeline:
- Next week for TMT labelling
- First week of August for sending me a dataset
Page 46: 2016-07-21. Reference samples for mapping index; Hsp modulation and thermal niche paper.
From SHC:
- FMU4 (ApGxL-03A)
- WP9 (ApGxL-11A)
- BRF4 (ApGxL-16A)
- SEB9 (ApGxL-22C)
- MB6 (ApGxL-26E)
Page 47: 2016-07-26. Learning mixed effects stat models
Mixed effects stat models let you include random or fixed variables, implemented in (lme4 package](http://lme4.r-forge.r-project.org/lMMwR/lrgprt.pdf). The difference? Summarized here in dynamic ecology blog.
As I understand it:
(Using sites as an example...)
Fixed effect...
- variable you're interested in
- continuous or categorical
- estimates values at each site, so if you have a lot of sites, it'll use more degrees of freedom
- syntax: (y~x+s)
Random effect...
- variable you want to control (blocking)
- categorical/discrete (Can not have continuous variable as a random effect))
- estimates variance among all sites, conserves degrees of freedom (also cant calculate p values)
- syntax: (yx,random=1|s )
- rule of thumb: sites should have roughly >5 levels ( 5 sites)
- comment in blog post says you can think of RE as groups having different slopes and or intercepts
Typing this out seems to make more sense. Now to go over some of the syntax....
This tutorial gives a good explanation.
It's hard to get p-values from mixed effects models, so one strategy is to make a full and null model with and without the variable of interest and running an anova. Don't use REML when doing these comparisons.
More syntax...
politeness.model = lmer(frequency ~ attitude + gender + (1|subject) + (1|scenario), data=politeness)This syntax (1|variable) specifies subject and scenario as random effects. It is a random intercept model.
This specifies a random slope model:
politeness.model = lmer(frequency ~ attitude + gender + (1+attitude|subject) + (1+attitude|scenario),data=politeness,REML=FALSE)This allows subjects and items to have difference slops and intercepts. Only thing changed is the random effect
Best practice to fit random slopes and intercepts! (Grueber et al. 2011, Journal of Evolutionary Biology; and the tutorial advocates for this because it reduces type I and II errors)
Notes, assumptions similar to fixed effects models
- Check for collinearity and influential data points
- check residuals, Q-Qplots
- One of the main shifts from linear models to mixed effect models was to account for non-independence (measuring outcome of same individual)
random effects note
So, a random effect is generally something that can be expected to have a nonsystematic, idiosyncratic, unpredictable, or “random” influence on your data. In experiments, that’s often “subject” and “item”, and you generally want to generalize over the idiosyncrasies of individual subjects and items.
fixed effects note
Fixed effects on the other hand are expected to have a systematic and predictable influence on your data.
Writing this up in a methods section
We used R (R Core Team, 2012) and lme4 (Bates, Maechler & Bolker, 2012) to perform a linear mixed effects analysis of the relationship between pitch and politeness. As fixed effects, we entered politeness and gender (without interaction term) into the model. As random effects, we had intercepts for subjects and items, as well as by-subject and by-item random slopes for the effect of politeness. Visual inspection of residual plots did not reveal any obvious deviations from homoscedasticity or normality. P-values were obtained by likelihood ratio tests of the full model with the effect in question against the model without the effect in question.
Page 48: 2016-07-27. Meeting with Steve Keller to discuss post doc idea (started here: Page 37: 2016-06-11. Quantifying natural selection in natural populations )
Raw notes from notebook:
Page 1
 Page 2
Page 2

Thoughts+ retyping notes:
- One challenge Steve brought up was that photoperiod is diff across lat and is not changing with climate. So when scientists do recipricol transplants between north and south populations, photoperiod is a confounding effect with temperature/climate.
- Selection gradients may not be increasing with climate if there is insufficient genetic variation to respond to selection. It could decrease. I need to think more carefully about how to connect selection gradients with population level dynamics. (I still need to read Ruth Shaw's aster modeling papers).
- RIght now, as I've pitched it, I have no manipulations which is something I need to determine whether temperature is actually increasing selection gradients.
- start cohorts at different times to control development
- for biotic interactions, manipulate floral display for pollintors
- induce herbivory- plant stress responses
- I could estimate kinship matrix in natural populations with many markers(thousands) and apply quantitative genetics techniques to identify constraints between different traits.
Post doc grants:
- Plant genome fellowship due in november (focused on crop or crop related plants)
- systems: sunflower, grasses, medacago, poplar(viability selection, high early life stage mortality), willows
- Fullbright for international opportunities
Page 49:2016-07-28. Quantitative genetics and the molecular basis of complex traits
Molecular biologists and quantitative genetics are intersted in,at some level, the molecular basis of complex traits. However, each field uses different approaches to this problem. Traditionally, the molecular biologist will manipulate a gene within a single genotype to observe its effect on a phenotype. On the other hand, a quantitative geneticist will take many different genotypes, shuffle genes around by mating individuals with each other, and then statistically assign the effect of genotypes in general on a phenotype.
It'd be interesting to merge both approaches: Knock out or in a gene for many genotypes within a mating design. This way, you can observe the effect on a particular gene within many different genotypes. Just a thought!
Paaby lab is doing a bit of this. She gave a talk earlier this year? Anyway, she picked a well known developmental pathway in worms(C. elegans) and used RNAi for many different species(I think) for a panel of genes.
Page 50: 2016-08-02. Picking a plant system for post doc idea
I plan on applying for Plant Genome Research Program (PGRP). Previous awards. I need a plany with a sequenced genome which is a crop or crop-related. List of sequenced genomes, list of genomes with "good" annotations
Mimulus guttatus (Monkey Flower)
Cool paper showing that there are annuals and perrenials which vary in morphology under a common garden. So, I could compare selection gradients for annuals vs perrenials.
- Read this lab's papers because they are interseted in similar things.
Leavenworthia alabamica
Annual secluded to Alabama and it has low population size. Populations/individuals vary in their reproductive mode: self compatible, self incompatible. So it'd be interesting to see how selection acts on these different reproductive forms.
Papers:
- Herman & Schoen 2016 and this one
- Secondary loss in self incompat
- [Compares selection gradients of self compt and self incompt plants] (http://www.amjbot.org/content/99/3/488.full)
Panicum virgatum (switch grass)
Perrenial with wide distribution from Canada to Mexico. We could look at episodic selection under a common garden across latitude. If you have performance on the y axis and x axis is lat(climate), and we have a mating design, we can analyze the data as function-valued traits. Growth would be a good option.
Measuring physiology: IR gas exchange analyzer
Measures photosynthetic rate and transpiration rate!
Cool technique to QTL with function valued traits here
Page 51: 2016-08-02; 2016-08-03. Climate cascade meeting
- Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
Multiple stressors ms: NJG gave me edits 2016-08-02, rework, then send to Sara. Aiming to submit next week?
Range limits ms: Go over figures, meet with NJG 2016-08-03 to go over intro, methods, and results.
Figure suggestions:
- recolor map, keep maps consistent
- shift cold tolerance vs tmin legend from horiztonal to vertical.
- double checkt he interaction of tmin and pre treatment temp; the betas
- create 2 panel fig for basal cold tolerance and hardening.
- recolor map, keep maps consistent
Thermal niche ms: Lacey's hands
HSP modulation paper: SHC's hands
Stressed in nature MS: Curtis' hands ; he was suppose to give me a timeline
Genome sequencing? Mlau's hands
Phylogenomics of common forest ants: SHC and Bernice assembling data matrix. ADN needs to send vouchers to Bernice.
- Ask about post doc (NJG and SHC think its ok to stay at same institution)
- Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper? Apply for funding? Suitor Travel Grant Deadline is october 31
- Biolunch: Should I talk about github?( SHC and NJG are ok with this but I need to think about my delivery and what people can "handle") Range limits? Dissertation talk (I want to give this in the Spring ( SHC says yes ))?
Page 52: 2016-08-04. Following up stats, range limits project
analysis of data with pre treatment temperature as continuous within an anova
## anova modelk.dat$pretreat_Temp<-as.numeric(as.character(k.dat$pretreat_Temp))cold.mod1<-aov(treatment_recovery_s~Tmin*pretreat_Temp+Colony,data=k.dat) # testing interaction between pre-treat temp and T min (both continuous)Df Sum Sq Mean Sq F value Pr(>F) Tmin 1 116145 116145 5.755 0.018765 * pretreat_Temp 1 261310 261310 12.949 0.000553 ***Tmin:pretreat_Temp 1 162568 162568 8.056 0.005747 ** Residuals 80 1614444 20181 analysis of data with pre treatment temperature as a factor within a linear model
##analysis of data with pre treatment temperature as a factor within a linear model
k.dat$pretreat_Temp<-as.factor(as.character(k.dat$pretreat_Temp))
cold.mod1<-lm(treatment_recovery_s~Tmin*pretreat_Temp+Colony,data=k.dat) #testing interaction between factors of pretreatment with Tmin(continuous)
#summary(cold.mod1)
#stepwise aic
qc<-stepAIC(cold.mod1,direction="both")
summary(qc)
#output:
summary(qc)
Call:
lm(formula = treatment_recovery_s ~ Tmin + pretreat_Temp + Tmin:pretreat_Temp,
data = k.dat)
Residuals:
Min 1Q Median 3Q Max
-292.69 -79.96 -10.13 69.04 355.98
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 210.58 363.71 0.579 0.56432
Tmin -24.64 24.27 -1.015 0.31321
pretreat_Temp0 450.14 514.37 0.875 0.38426
pretreat_Temp25 1796.59 514.37 3.493 0.00080 ***
pretreat_Temp5 1173.92 514.37 2.282 0.02527 *
Tmin:pretreat_Temp0 40.73 34.33 1.186 0.23916
Tmin:pretreat_Temp25 114.57 34.33 3.338 0.00131 **
Tmin:pretreat_Temp5 76.71 34.33 2.235 0.02837 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 124 on 76 degrees of freedom
Multiple R-squared: 0.4577, Adjusted R-squared: 0.4078
F-statistic: 9.164 on 7 and 76 DF, p-value: 3.644e-08
More digestable table:
knitr::kable(summary(qc)$coefficients)| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 210.58099 | 363.71495 | 0.5789726 | 0.5643197 |
| Tmin | -24.64324 | 24.27295 | -1.0152553 | 0.3132054 |
| pretreat_Temp0 | 450.14412 | 514.37061 | 0.8751358 | 0.3842574 |
| pretreat_Temp25 | 1796.59479 | 514.37061 | 3.4928022 | 0.0008002 |
| pretreat_Temp5 | 1173.91549 | 514.37061 | 2.2822367 | 0.0252738 |
| Tmin:pretreat_Temp0 | 40.72533 | 34.32714 | 1.1863889 | 0.2391643 |
| Tmin:pretreat_Temp25 | 114.57348 | 34.32714 | 3.3376940 | 0.0013101 |
| Tmin:pretreat_Temp5 | 76.71280 | 34.32714 | 2.2347566 | 0.0283715 |
Hardening ability
cold.mod8<-aov(hardening~Tmin*PT+Colony,data=mew6)qc8<-stepAIC(cold.mod8,direction="both")summary(qc8) Df Sum Sq Mean Sq F value Pr(>F) Tmin 1 85850 85850 5.903 0.02055 * PT 2 550143 275071 18.915 3.01e-06 ***Colony 17 1435781 84458 5.808 6.88e-06 ***Tmin:PT 2 179795 89897 6.182 0.00513 ** Residuals 34 494455 14543 Good post to read for understanding interactions here
2016-08-11 updated analyses
Basal cold tolerance re-analyzed
df SS MS F-value P-value Tmin 1 114575 114575 6.757 0.0122 Pre-treatment 3 623523 207841 12.257 <0.001 Tmin × Pre-treatment 3 189451 63150 3.724 0.0169 Colony 17 228419 13436 0.792 0.6931 Residuals 51 864771 16956
Cold hardening re-analyzed (double checked)
df SS MS F-value P-value Tmin 1 411796 411796 26.318 <0.001 Pre-treatment 2 363498 181749 11.616 <0.001 Tmin × Pre-treatment 2 98308 49154 3.141 0.055986 Colony 17 1285635 75626 4.833 <0.001 Residuals 34 531992 15647
Interaction non-significant; the change was caused by a mistake made by consolidating scripts.
Page 53: 2016-08-08. Post doc ideas part 2
1. How does selection operate on the life histories of poplar? [Similar to this post doc listing] (http://evol.mcmaster.ca/~brian/evoldir/PostDocs/INRAFrance.EvolQuantGenetics)
Approach: Identify and characterize how natural selectin operates at different life stages of poplar
- Measure selection gradients on age structured populations in the field
- Is it possible to heat shock leaves out in the field?
- Viability selection( Mojica & Kelly ref) : One thing missing from selection studies is that organisms can die before expressing a trait (They confusingly call this the invisible fraction of variation). Can we test this by taking cuttings and planting them? Or does it have to be from seeds? (I think the latter)
- Good natural history
How does contemporary episodes of natural selection compare with past local adaptation to climate?
Approach: Compare selection in the field to common garden. There is a cool paper by Kingsolver et al. 2012 that sugguests we account for environmental covariation with selection gradient analyses. If we have a relatedness matrix, we can see if individuals are spatially clustered with environment.
2. How does selection operate on populations of monkeyflowers (Mimulus guttatus) with different modes of reproduction?
Approach: Identify and characterize how natural selection operates on perennials and annuls Which one is more susceptbile? Are there shifts between one or the other?
- Measure selection gradients across a whole cline (whole west coast of US) for perrenials and annuals.
- Perrenials experience greater within generation variation than among--so they may harbor greater plasticity than annuals.
3. Gladecresses Leavenworthia alabamica
- measure selection gradients between self compatible vs self incompatible for populations in Alabama.
- Low adaptive potential in self compatible vs self incompatible.
4. Identifying specific genotypes for optimal growth in the Shrub willow (Salix pupurea)
Approach1: Evaluate growth as a function valued trait across latitudinal cline
- Mate and plot genotypes in the field. Or take clippings and plant?
- Measure growth across latitude.
Approach2: Evaluate growth as a function valued trait within a common garden
- Possible to have them reared at 6 temperatures and 3 moisture levels? *
Analysis: Determine shifts in growth reaction norms.
Page 54: 2016-08-10. Climate cascade meeting
- Project updates:
- Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
- Multiple stressors ms: SHC's hands- discussion is too disjointed, reworking organization
- Range limits ms: Fixed figures, go over!
- Thermal niche ms: Lacy and I are working on it. Discussion left to do
- HSP modulation paper: SHC's hands
- Stressed in nature MS: Curtis' hands ; he was suppose to give me a timeline
- Genome sequencing? Mlau's hands
- Phylogenomics of common forest ants: ADN to send Bernice samples this week.
- Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Biolunch, working title: Strategies for achieving reproducible research ; get picture of the meeting
Page 55: 2016-08-11. Overlaying raster files in a map in R
Good link to show how to overlay here. I've had to use this to plot climate cut offs (example: here)
Some code:
Cropping world map, I set coords to region I'm interested in: Maine
w2 <- getData('worldclim', var='bio', res=.5,lat=45,lon=-68) # grab worldclim data; with .5 res you need to specify coordinatesextent<-c(-72,-65,42,48)bew<-crop(w2,extent)Here is the code to make cut offs: designate extreme values and then plotting it will be easy
You have to get rid of NAs and assign to variable.
Tm<-na.omit(bew[[5]])Tm[bew[[5]] < 246.5] <- 100 # absentTm[bew[[5]] > 246.5] <- 1Here is plotting the cut off
dbio2$coco<-ifelse(dbio2$Found_Notfound=="1","red","black") # specify color of points base don presence absenceplot(lar[[5]],col=c("white","grey75"),legend=F)map("worldHires",c("USA","Canada"),add=TRUE)map("state", c('maine','vermont','new hampshire'), add = TRUE)points(dbio2$Lon,dbio2$Lat,pch=16,col=dbio2$coco)Page 56: 2016-08-16 range limits paper, data analysis of chill coma recovery time (CCRT) revisited
From my G matrix anlaysis, I find variation in the cooler-warmer axis. So for my statistics for relating CCRT to local environment (to see if they're locally adapted), I used an ANCOVA:
CCRT~ pre-treatment temp * Tmin This just says whether the relationship between CCRT and Tmin at each pre-treatment temperature are different or not. But what I may want, is an estimate of those relationships. So I should run a regression or mixed effect model to generalize to the whole population.
Mixed effect model with pretreatment * Tmin interaction, random intercept and slope? for every colony measured at each pretreatment temp
mod5.r<-lmer(formula=inv_c~pretreat_Temp*Tmin+(1+pretreat_Temp|Colony),REML=TRUE,data=test)I'll compare this model to:
Mixed effect model with fixed effect of Tmin, random intercept and slope? for every colony measured at each pretreatment temp
mod3<-lmer(formula=inv_c~Tmin+(1+pretreat_Temp|Colony),REML=TRUE,data=test)and also compare it to:
Mixed effect model with fixed effect of Tmin and pretreatment temp, random intercept and slope? for every colony measured at each pretreatment temp
mod4<-lmer(formula=inv_c~pretreat_Temp+Tmin+(1+pretreat_Temp|Colony),REML=TRUE,data=test) my "comparison" using anova function:
refitting model(s) with ML (instead of REML)Data: testModels:mod3: inv_c ~ Tmin + (1 + pretreat_Temp | Colony)mod2: inv_c ~ pretreat_Temp + (1 + pretreat_Temp | Colony)mod4: inv_c ~ pretreat_Temp + Tmin + (1 + pretreat_Temp | Colony)mod5.r: inv_c ~ pretreat_Temp * Tmin + (1 + pretreat_Temp | Colony) Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq) mod3 13 555.36 602.00 -264.68 529.36 mod2 15 544.10 597.91 -257.05 514.10 15.2606 2 0.0004855 ***mod4 16 543.62 601.02 -255.81 511.62 2.4798 1 0.1153190 mod5.r 19 540.10 608.26 -251.05 502.10 9.5188 3 0.0231317 * ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 mod5.r is stat diff from the other more simple models
Let's look at the output:
Linear mixed model fit by REML ['lmerMod']Formula: inv_c ~ pretreat_Temp * Tmin + (1 + pretreat_Temp | Colony) Data: testREML criterion at convergence: 525.8Scaled residuals: Min 1Q Median 3Q Max -1.9347 -0.5625 -0.1789 0.4116 5.4326 Random effects: Groups Name Variance Std.Dev. Corr Colony (Intercept) 0.03646 0.1909 pretreat_Temp0 0.15330 0.3915 -0.23 pretreat_Temp25 0.20398 0.4516 -0.92 -0.13 pretreat_Temp5 0.26667 0.5164 -0.17 -0.50 0.50 Residual 0.32402 0.5692 Number of obs: 267, groups: Colony, 18Fixed effects: Estimate Std. Error t value(Intercept) 3.59188 1.03188 3.481pretreat_Temp0 -2.90454 1.68322 -1.726pretreat_Temp25 -4.39184 1.80599 -2.432pretreat_Temp5 -4.47550 1.96022 -2.283Tmin 0.11598 0.06922 1.675pretreat_Temp0:Tmin -0.23723 0.11283 -2.102pretreat_Temp25:Tmin -0.28354 0.12088 -2.346pretreat_Temp5:Tmin -0.30516 0.13104 -2.329Correlation of Fixed Effects: (Intr) prt_T0 pr_T25 prt_T5 Tmin p_T0:T p_T25:pretrt_Tmp0 -0.519 prtrt_Tmp25 -0.770 0.183 pretrt_Tmp5 -0.443 -0.039 0.499 Tmin 0.997 -0.517 -0.766 -0.442 prtrt_Tm0:T -0.518 0.997 0.184 -0.037 -0.520 prtrt_T25:T -0.768 0.184 0.997 0.498 -0.770 0.185 prtrt_Tm5:T -0.443 -0.037 0.498 0.997 -0.445 -0.036 0.500Considering only the random effect of colony
mod2<-lmer(formula=treatment_recovery_s.x~pretreat_Temp+(1|Colony),REML=TRUE,data=test)mod3<-lmer(formula=treatment_recovery_s.x~Tmin+(1|Colony),REML=TRUE,data=test)mod4<-lmer(formula=treatment_recovery_s.x~pretreat_Temp+Tmin+(1+pretreat_Temp|Colony),REML=TRUE,data=test)#mod5.r<-lmer(formula=inv_c~pretreat_Temp*Tmin+(1|Colony),REML=TRUE,data=test)mod6<-lmer(formula=treatment_recovery_s.x~pretreat_Temp*Tmin+(1|Colony),REML=TRUE,data=test)anova(mod3,mod4,mod2,mod6)mod3: treatment_recovery_s.x ~ Tmin + (1 | Colony)mod2: treatment_recovery_s.x ~ pretreat_Temp + (1 | Colony)mod4: treatment_recovery_s.x ~ pretreat_Temp + Tmin + (1 | Colony)mod6: treatment_recovery_s.x ~ pretreat_Temp * Tmin + (1 | Colony) Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq) mod3 4 3628.0 3642.4 -1810.0 3620.0 mod2 6 3583.1 3604.6 -1785.5 3571.1 48.9531 2 2.344e-11 ***mod4 7 3577.2 3602.3 -1781.6 3563.2 7.8337 1 0.005128 ** mod6 10 3564.4 3600.2 -1772.2 3544.4 18.8832 3 0.000289 ***---model output for mod6
Linear mixed model fit by REML ['lmerMod']Formula: treatment_recovery_s.x ~ pretreat_Temp * Tmin + (1 | Colony) Data: testREML criterion at convergence: 3649.7Scaled residuals: Min 1Q Median 3Q Max -3.2557 -0.6656 -0.1116 0.4587 3.8248 Random effects: Groups Name Variance Std.Dev. Colony (Intercept) 752.9 27.44 Residual 33965.8 184.30 Number of obs: 280, groups: Colony, 19Fixed effects: Estimate Std. Error t value(Intercept) 207.92 291.37 0.714pretreat_Temp0 439.58 396.48 1.109pretreat_Temp25 1736.31 395.31 4.392pretreat_Temp5 1215.86 399.33 3.045Tmin -24.52 19.57 -1.253pretreat_Temp0:Tmin 39.34 26.65 1.476pretreat_Temp25:Tmin 109.35 26.52 4.124pretreat_Temp5:Tmin 79.62 26.73 2.979Correlation of Fixed Effects: (Intr) prt_T0 pr_T25 prt_T5 Tmin p_T0:T p_T25:pretrt_Tmp0 -0.678 prtrt_Tmp25 -0.681 0.500 pretrt_Tmp5 -0.674 0.495 0.497 Tmin 0.997 -0.676 -0.679 -0.672 prtrt_Tm0:T -0.676 0.997 0.498 0.493 -0.678 prtrt_T25:T -0.680 0.499 0.997 0.496 -0.682 0.501 prtrt_Tm5:T -0.674 0.496 0.497 0.997 -0.677 0.497 0.499Page 57: 2016-08-25. Hsp modulation follow up stats
summary(aov(log10(B_40)~axis3_desig,data=mergy))
Df Sum Sq Mean Sq F value Pr(>F)
axis3_desig 3 4.947 1.6490 7.154 0.000413 ***
Residuals 52 11.986 0.2305
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
I separated out groupings based on phylogenetic axes. The model anova is significant.
Now I'll do a post hoc test.
TukeyHSD(aov(log10(B_40)~axis3_desig,data=mergy))
diff lwr upr p adj
North-A. picea 0.1185330 -0.3739818 0.6110478 0.9189644
South-A. picea -1.0921848 -1.7596714 -0.4246982 0.0003710
zAxis 2 A. picea-A. picea 0.2516912 -0.5104439 1.0138263 0.8169654
South-North -1.2107178 -1.9910398 -0.4303958 0.0007709
zAxis 2 A. picea-North 0.1331582 -0.7295202 0.9958366 0.9765503
zAxis 2 A. picea-South 1.3438760 0.3706435 2.3171085 0.0031663Page 58: 2016-08-29 & 30. Climate cascade meeting
- Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
Multiple stressors ms: working on SHC edits
- Send out Wednesday.
Range limits ms: Go over figure; SHC has ms; eta? Not looked at it.
- sampling map: make larger, points should be gray; sites that were used for common garden should have a gold outline
- fig 6, cold phys; get rid of "cold", use different words.
Thermal niche ms: Lacey and I working on discussion
HSP modulation paper: SHC submitted
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- update: Curtis can no longer work+ write on project
The DF 20140717 sample box was found when we dug through all the freezers in the winter and I didn't have time to extract RNA and qPCR them all. The HF 20140812 box was the box we weren't able to find anywhere.
There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- Genome sequencing? Mlau's hands
- Phylogenomics of common forest ants: status?
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- construct talk; when to give practice talk ?
Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing and then get everything signed.
- Wrote up suiter award app. I need to find out pricing and then get everything signed.
Biolunch, working title: Strategies for achieving reproducible research Sept 2nd.
Page 59: 2016-09-01. SHC lab meeting Fall 2016
| Room | Date | Activity | Person.in.Charge | Breakfast |
|---|---|---|---|---|
| 124 | Sept. 8 | IDPs | Sara | Sara |
| 124 | Sept. 15 | American Naturalist paper | Sara | Megna |
| 122 | Sept. 22 | Experimental design | Megna | Katie |
| 124 | Sept. 29 | Manuscript - A. picea range limits | Andrew | Laurel |
| 124 | Oct. 6 | Proposal - NSF post-doc fellowship | Andrew | Delaney |
| 124 | Oct. 13 | Experimental design | Julia | Julia |
| 122 | Oct. 20 | Research update | Bonnie | Bonnie |
| 124 | Oct. 27 | Results presentation | Delaney | Delaney |
| 124 | Nov. 3 | Paper discussion | Laurel | Sara |
| 124 | Nov. 10 | Results discussion | Laurel | Laurel |
| 122 | Nov. 17 | Manuscript - CNP in Aphaenogaster | Katie | Bonnie |
| NA | 24-Nov | Thanksgiving | ||
| 124 | Dec. 1 | Meeting talk - range limits | Andrew | Sara |
| 124 | Dec. 8 | Dimensions of Biodiversity new papers!!! | Everyone | Andrew |
Note dietary requirements for breakfasts:
- Dairy-free options
- No coconut
- No nuts in baked goods
- No honeydew melon
Including notes from meeting (added 2016-09-02)
- LSO needs to check monthly eye wash, chemical inventory, lab safety
- Do your lab safety training.
-
Tuesday morning (2016-09-06): Schedule time to look for ants, collect ~ 20.
2016-09-01: Paper notes: Paccard et al. 2016
ref: Paccard A, Van Buskirk J, Willi Y, Eckert CG, Bronstein JL. 2016. Quantitative Genetic Architecture at Latitudinal Range Boundaries: Reduced Variation but Higher Trait Independence. The American Naturalist.
Quick and dirty: They compared variance-covariance G matrices among 9 populations in a Arabdopsis species that spans a cline. It was in a common garden with 2 levels of moisture treatments.
Findings:
- Genetic variance washighest at the middle of their range and lowest at the edges (south and north)
- More trait indepdence at the northern part of their range
Making sense of the properties of G
Confusing sentence in methods: *We calculated four measures of multivariate evolutionary
potential and G-matrix geometry (size, sphericity, and orientation)
for each treatment and population.*
Separate or the same item?
- Size: sum of genetic variances across all traits. I guess this means the total amount of genetic variance.
- Sphericity = # of dimensions: Sum of all eigenvalues / first eigenvalue. It tells you how independent traits are. If it is 1 then gmax or the first pc explains most of the variation. But if it is a high number (# of dimensions of G), then it tells you many traits are independent and variances are distributed among traits. It can also tell you whether genetic constraints exist in certain directions without specifying direction
- Orientation of G relative to common standard vector: Compare dmax (dominant eigenvector of variance-covariance matrix of population means for 10 traits across the 9 populations--- D matrix describes population divergence ). For each population they meaured the orientation as the absolute value of the angle between dmax and gmax.
- Response to selection: Random skewers method: THey calculate change in phenotype by simulating Betas in the delta Z = G * Beta
Page 61: 2016-09-06. Playing with rpart with range limit data
Using bioclim variables to classify presence-absence
Guidance for picking "best" tree
- Convention is to pick one with the lowest cross-validate relative error or smallest(simplest) tree within 1 standard error of best tree
-
Full dataset layout
str(dbio2)'data.frame': 102 obs. of 38 variables: $ n : int 1 2 3 4 5 6 7 8 9 10 ... $ date : int 19960507 20140709 20140709 20140710 20050625 20030715 20050625 20130718 19910901 20050630 ... $ state : Factor w/ 1 level "Maine": 1 1 1 1 1 1 1 1 1 1 ... $ county : Factor w/ 23 levels "","cumberland",..: 23 2 8 8 6 6 6 21 7 6 ... $ locality : Factor w/ 84 levels "","18-LP-4C",..: 81 42 17 17 6 3 4 76 61 67 ... $ habitat : Factor w/ 12 levels ""," ","Behind dining hall",..: 11 8 5 6 NA NA NA 3 12 NA ... $ Lat : num 43.6 43.9 43.9 43.9 44.3 ... $ Lon : num -70.8 -70.2 -69.7 -69.7 -68.3 ... $ masl : num 158 NA NA NA 68 100 230 NA NA 105 ... $ subfamily : Factor w/ 2 levels "","Myrmicinae": 2 2 2 2 2 2 2 2 2 2 ... $ ant.genus : Factor w/ 2 levels "","Aphaenogaster": 2 2 2 2 2 2 2 2 2 2 ... $ ant.species : Factor w/ 2 levels "","picea": 2 2 2 2 2 2 2 2 2 2 ... $ code : Factor w/ 2 levels "","aphpic": 2 2 2 2 2 2 2 2 2 2 ... $ collection : Factor w/ 75 levels "","Aaron","AcadiaNP",..: 5 1 4 1 3 3 3 1 6 7 ... $ collector : Factor w/ 11 levels "Aaron","Acadia BioBlitz",..: 10 3 3 3 8 2 8 4 10 9 ... $ Found_Notfound: int 1 1 1 1 1 1 1 1 1 1 ... $ MAT : num 7 7.6 7.8 7.8 6.9 6.6 6.3 6.6 6.6 6.8 ... $ MDR : num 129 108 105 105 107 107 106 109 124 110 ... $ ISO : num 32 28 28 28 28 28 28 29 30 28 ... $ SD : num 94.2 92.7 90.5 90.5 90.7 ... $ Tmax : num 27.1 26.3 26 26 25.5 25.2 24.8 24.9 27.1 25.9 ... $ Tmin : num -132 -115 -107 -107 -117 -121 -123 -120 -142 -125 ... $ TAR : num 403 378 367 367 372 373 371 369 413 384 ... $ TWQ : num 24 33 37 37 -22 -25 -28 -23 20 25 ... $ TDQ : num 179 186 192 192 183 180 177 177 -53 186 ... $ TwarmQ : num 188 193 192 192 183 180 177 177 189 186 ... $ TminQ : num -57 -47 -42 -42 -52 -56 -59 -54 -66 -58 ... $ AP : num 1195 1146 1157 1157 1261 ... $ PWM : num 131 123 125 125 144 148 150 140 110 127 ... $ PDM : num 86 79 76 76 78 79 81 77 69 81 ... $ PSD : num 12 13 14 14 17 18 18 17 11 14 ... $ PWQ : num 349 335 341 341 388 401 407 385 301 343 ... $ PDQ : num 267 244 244 244 245 250 256 245 231 248 ... $ PwarmQ : num 275 248 244 244 245 250 256 245 268 248 ... $ PminQ : num 293 293 297 297 342 354 359 340 240 294 ... $ var : Factor w/ 2 levels "absent","present": 2 2 2 2 2 2 2 2 2 2 ... $ color : chr "red" "red" "red" "red" ... $ coco : chr "red" "red" "red" "red" ...All bioclim variables
knitr::kable(round(cor(dbio2[17:35]),3))
| MAT | MDR | ISO | SD | Tmax | Tmin | TAR | TWQ | TDQ | TwarmQ | TminQ | AP | PWM | PDM | PSD | PWQ | PDQ | PwarmQ | PminQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAT | 1.000 | -0.273 | 0.352 | -0.637 | 0.663 | 0.876 | -0.512 | -0.740 | 0.620 | 0.852 | 0.948 | 0.560 | 0.598 | 0.769 | -0.265 | 0.495 | 0.793 | -0.878 | 0.684 |
| MDR | -0.273 | 1.000 | 0.541 | 0.787 | 0.483 | -0.674 | 0.913 | 0.137 | -0.722 | 0.168 | -0.519 | -0.606 | -0.647 | -0.437 | -0.587 | -0.733 | -0.399 | 0.525 | -0.678 |
| ISO | 0.352 | 0.541 | 1.000 | -0.047 | 0.537 | 0.104 | 0.179 | -0.387 | -0.027 | 0.402 | 0.249 | 0.119 | 0.060 | 0.218 | -0.470 | -0.040 | 0.303 | -0.127 | 0.072 |
| SD | -0.637 | 0.787 | -0.047 | 1.000 | 0.133 | -0.916 | 0.967 | 0.526 | -0.859 | -0.143 | -0.848 | -0.836 | -0.843 | -0.717 | -0.341 | -0.861 | -0.724 | 0.771 | -0.898 |
| Tmax | 0.663 | 0.483 | 0.537 | 0.133 | 1.000 | 0.229 | 0.299 | -0.506 | -0.031 | 0.939 | 0.398 | -0.056 | -0.015 | 0.344 | -0.700 | -0.180 | 0.364 | -0.395 | 0.041 |
| Tmin | 0.876 | -0.674 | 0.104 | -0.916 | 0.229 | 1.000 | -0.860 | -0.649 | 0.845 | 0.511 | 0.980 | 0.771 | 0.809 | 0.818 | 0.106 | 0.775 | 0.826 | -0.916 | 0.884 |
| TAR | -0.512 | 0.913 | 0.179 | 0.967 | 0.299 | -0.860 | 1.000 | 0.371 | -0.844 | -0.009 | -0.752 | -0.785 | -0.800 | -0.622 | -0.471 | -0.854 | -0.619 | 0.692 | -0.844 |
| TWQ | -0.740 | 0.137 | -0.387 | 0.526 | -0.506 | -0.649 | 0.371 | 1.000 | -0.452 | -0.589 | -0.714 | -0.577 | -0.586 | -0.721 | 0.258 | -0.466 | -0.712 | 0.740 | -0.671 |
| TDQ | 0.620 | -0.722 | -0.027 | -0.859 | -0.031 | 0.845 | -0.844 | -0.452 | 1.000 | 0.232 | 0.783 | 0.806 | 0.847 | 0.741 | 0.401 | 0.848 | 0.709 | -0.756 | 0.878 |
| TwarmQ | 0.852 | 0.168 | 0.402 | -0.143 | 0.939 | 0.511 | -0.009 | -0.589 | 0.232 | 1.000 | 0.646 | 0.168 | 0.218 | 0.526 | -0.561 | 0.070 | 0.548 | -0.613 | 0.285 |
| TminQ | 0.948 | -0.519 | 0.249 | -0.848 | 0.398 | 0.980 | -0.752 | -0.714 | 0.783 | 0.646 | 1.000 | 0.730 | 0.760 | 0.824 | -0.037 | 0.697 | 0.845 | -0.918 | 0.840 |
| AP | 0.560 | -0.606 | 0.119 | -0.836 | -0.056 | 0.771 | -0.785 | -0.577 | 0.806 | 0.168 | 0.730 | 1.000 | 0.957 | 0.731 | 0.381 | 0.955 | 0.783 | -0.632 | 0.941 |
| PWM | 0.598 | -0.647 | 0.060 | -0.843 | -0.015 | 0.809 | -0.800 | -0.586 | 0.847 | 0.218 | 0.760 | 0.957 | 1.000 | 0.799 | 0.424 | 0.976 | 0.799 | -0.733 | 0.971 |
| PDM | 0.769 | -0.437 | 0.218 | -0.717 | 0.344 | 0.818 | -0.622 | -0.721 | 0.741 | 0.526 | 0.824 | 0.731 | 0.799 | 1.000 | -0.117 | 0.695 | 0.966 | -0.856 | 0.856 |
| PSD | -0.265 | -0.587 | -0.470 | -0.341 | -0.700 | 0.106 | -0.471 | 0.258 | 0.401 | -0.561 | -0.037 | 0.381 | 0.424 | -0.117 | 1.000 | 0.560 | -0.157 | 0.031 | 0.310 |
| PWQ | 0.495 | -0.733 | -0.040 | -0.861 | -0.180 | 0.775 | -0.854 | -0.466 | 0.848 | 0.070 | 0.697 | 0.955 | 0.976 | 0.695 | 0.560 | 1.000 | 0.707 | -0.649 | 0.940 |
| PDQ | 0.793 | -0.399 | 0.303 | -0.724 | 0.364 | 0.826 | -0.619 | -0.712 | 0.709 | 0.548 | 0.845 | 0.783 | 0.799 | 0.966 | -0.157 | 0.707 | 1.000 | -0.803 | 0.847 |
| PwarmQ | -0.878 | 0.525 | -0.127 | 0.771 | -0.395 | -0.916 | 0.692 | 0.740 | -0.756 | -0.613 | -0.918 | -0.632 | -0.733 | -0.856 | 0.031 | -0.649 | -0.803 | 1.000 | -0.835 |
| PminQ | 0.684 | -0.678 | 0.072 | -0.898 | 0.041 | 0.884 | -0.844 | -0.671 | 0.878 | 0.285 | 0.840 | 0.941 | 0.971 | 0.856 | 0.310 | 0.940 | 0.847 | -0.835 | 1.000 |
rpart predictive model: full bioclim
vars<-as.data.frame(cbind(dbio2[,17:35],V1=dbio2[,36])) #all bioclim variablesform<-as.formula(V1~.)tree.1<-rpart(form,data=vars,control=rpart.control(minsplit=20,cp=0),method="class")printcp(tree.1)plotcp(tree.1)rpart.plot(tree.1,type=3,extra=100)classification tree
Table statistics of model:
| CP | nsplit | rel error | xerror | xstd |
|---|---|---|---|---|
| 0.42 | 0 | 1.00 | 1.26 | 0.0981595 |
| 0.12 | 1 | 0.58 | 0.82 | 0.0990346 |
| 0.06 | 2 | 0.46 | 0.76 | 0.0976589 |
| 0.00 | 5 | 0.28 | 0.66 | 0.0944956 |

model accuracy
m<-predict(tree.1,vars[-20])m.pre<-ifelse(m[,1]< m[,2],"present","absent")#confusion matrix#following this tutorial#http://eric.univ-lyon2.fr/~ricco/tanagra/fichiers/en_Tanagra_Validation_Croisee_Suite.pdfmc<-table(vars$V1,m.pre);mcsum(ifelse(vars$V1== m.pre,1,0))/nrow(vars)Confusion matrix indicating 86.2% accuracy:
| absent | present | |
|---|---|---|
| absent | 42 | 8 |
| present | 6 | 46 |
Subset of bioclim variables:
sub<-data.frame(cbind(dbio2$MAT,dbio2$Tmin,dbio2$SD,dbio2$TAR,dbio2$ISO,dbio2$MDR,dbio2$AP,dbio2[,31]))names(sub)<-c("MAT","Tmin","SD","TAR","ISO","MDR","AP","PSD")knitr::kable(round(cor(sub),3))| MAT | Tmin | SD | TAR | ISO | MDR | AP | PSD | |
|---|---|---|---|---|---|---|---|---|
| MAT | 1.000 | 0.876 | -0.637 | -0.512 | 0.352 | -0.273 | 0.560 | -0.265 |
| Tmin | 0.876 | 1.000 | -0.916 | -0.860 | 0.104 | -0.674 | 0.771 | 0.106 |
| SD | -0.637 | -0.916 | 1.000 | 0.967 | -0.047 | 0.787 | -0.836 | -0.341 |
| TAR | -0.512 | -0.860 | 0.967 | 1.000 | 0.179 | 0.913 | -0.785 | -0.471 |
| ISO | 0.352 | 0.104 | -0.047 | 0.179 | 1.000 | 0.541 | 0.119 | -0.470 |
| MDR | -0.273 | -0.674 | 0.787 | 0.913 | 0.541 | 1.000 | -0.606 | -0.587 |
| AP | 0.560 | 0.771 | -0.836 | -0.785 | 0.119 | -0.606 | 1.000 | 0.381 |
| PSD | -0.265 | 0.106 | -0.341 | -0.471 | -0.470 | -0.587 | 0.381 | 1.000 |
Classification tree with subset of bioclim
vars<-as.data.frame(cbind(sub,V1=dbio2[,36]))#names(vars)[1]<-"V1"form<-as.formula(V1~.)tree.1<-rpart(form,data=vars,control=rpart.control(minsplit=20,cp=0),method="class")printcp(tree.1)plotcp(tree.1)rpart.plot(tree.1,type=3,extra=100)output of classification tree
Table statistics of model:
| CP | nsplit | rel error | xerror | xstd |
|---|---|---|---|---|
| 0.38 | 0 | 1.00 | 1.24 | 0.0986179 |
| 0.14 | 1 | 0.62 | 1.00 | 0.1009756 |
| 0.04 | 2 | 0.48 | 0.84 | 0.0994100 |
| 0.02 | 6 | 0.32 | 0.66 | 0.0944956 |
| 0.00 | 7 | 0.30 | 0.52 | 0.0880285 |

model accuracy
m<-predict(tree.1,vars[-9])m.pre<-ifelse(m[,1]< m[,2],"present","absent")knitr::kable(mc)Confusion matrix indicating 85.2% accuracy
| absent | present | |
|---|---|---|
| absent | 46 | 4 |
| present | 11 | 41 |
Page 62: 2016-09-06. Climate cascade meeting
- Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
Multiple stressors ms:
- SHC hands
Range limits ms: Aaron made comments, go over with Nick
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- update: Curtis can no longer work+ write on project
There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- construct talk; when to give practice talk ?
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing and then get everything signed.
- construct talk; when to give practice talk ?
Notes: Only NJG and ANBE in attendance.
- Go over thesis layout next time
Page 63: 2016-09-07. PCA update for range limit data ; see * Page 63: 2016-09-07. PCA update for range limit data
Aaron wants to explore PCA decomposition of bioclim variables
PCA of all bioclim
nm<-princomp(scale(dbio2[,17:35]))knitr::kable(round(nm$loadings[,1:4],3))Table of loadings
| Comp.1 | Comp.2 | Comp.3 | Comp.4 | |
|---|---|---|---|---|
| MAT | 0.238 | -0.242 | 0.191 | -0.079 |
| MDR | -0.192 | -0.307 | -0.347 | 0.086 |
| ISO | 0.037 | -0.309 | -0.614 | -0.515 |
| SD | -0.267 | -0.124 | 0.000 | 0.393 |
| Tmax | 0.052 | -0.451 | 0.099 | 0.239 |
| Tmin | 0.281 | -0.026 | 0.184 | -0.206 |
| TAR | -0.248 | -0.211 | -0.129 | 0.327 |
| TWQ | -0.205 | 0.213 | 0.151 | -0.155 |
| TDQ | 0.259 | 0.111 | 0.034 | 0.002 |
| TwarmQ | 0.128 | -0.389 | 0.247 | 0.209 |
| TminQ | 0.274 | -0.112 | 0.140 | -0.205 |
| AP | 0.258 | 0.103 | -0.324 | 0.158 |
| PWM | 0.268 | 0.100 | -0.230 | 0.275 |
| PDM | 0.259 | -0.108 | -0.046 | 0.164 |
| PSD | 0.052 | 0.413 | -0.107 | 0.240 |
| PWQ | 0.256 | 0.180 | -0.215 | 0.198 |
| PDQ | 0.259 | -0.124 | -0.122 | 0.075 |
| PwarmQ | -0.263 | 0.107 | -0.228 | -0.014 |
| PminQ | 0.282 | 0.065 | -0.130 | 0.143 |
Screeplot of PCA of all bioclim vars

Variance explained
summary(nm)
Importance of components:
Comp.1 Comp.2 Comp.3 Comp.4 Comp.5
Standard deviation 3.4169139 2.0868333 1.00881816 0.72270248 0.71067369
Proportion of Variance 0.6205736 0.2314732 0.05409423 0.02776159 0.02684514
Cumulative Proportion 0.6205736 0.8520468 0.90614101 0.93390259 0.96074773PC1 explains 62%, PC2 explains 23%, PC3 explains 5%.
Statistical analysis: Using logistic regression, glm() function for first 3 PCs
dmo1<-glm(dbio2$Found_Notfound~pca.clim[,1]+pca.clim[,2]+pca.clim[,3],family="binomial")summary(dmo1) Call:glm(formula = dbio2$Found_Notfound ~ pca.clim[, 1] + pca.clim[, 2] + pca.clim[, 3], family = "binomial")Deviance Residuals: Min 1Q Median 3Q Max -1.6588 -0.9896 0.3712 0.9299 2.3119 Coefficients: Estimate Std. Error z value Pr(>|z|) (Intercept) -0.11715 0.24828 -0.472 0.63702 pca.clim[, 1] 0.23114 0.08479 2.726 0.00641 ** pca.clim[, 2] -0.57836 0.15037 -3.846 0.00012 ***pca.clim[, 3] -0.19877 0.24715 -0.804 0.42126 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1(Dispersion parameter for binomial family taken to be 1) Null deviance: 141.36 on 101 degrees of freedomResidual deviance: 112.62 on 98 degrees of freedomAIC: 120.62Number of Fisher Scoring iterations: 5#more digestable tableknitr::kable(round(summary(dmo1)$coefficients,3))Table output of logistic regression
| Estimate | Std. Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| (Intercept) | -0.117 | 0.248 | -0.472 | 0.637 |
| pc1 | 0.231 | 0.085 | 2.726 | ** 0.006** |
| pc2 | -0.578 | 0.150 | -3.846 | 0.000 |
| pc3 | -0.199 | 0.247 | -0.804 | 0.421 |
Overlaying presence-absence onto climate space as represented by PCs

Aaron's thoughts
Hi Andrew, The scree plot suggests both PC1 and maybe PC2, not definitely not PC3 are useful. The GLM supports this. The loadings on PC2 are clear: MDR, ISO, Tmax, TwarmQ, PSD, none of which load heavily on PC1 But the loadings on PC1 are a mess. None exceed 0.3 in loading, and the 0.2-0.3 (absolute values) are: MAT, SD, Tmin, TAR, TDQ, TminQ, AP, PWM, PDM, PDQ, PWarmQ, and PminQ. Looks to me like a lot of min temps and precip on PC1 and maxima on PC2, but I don’t know my bioclim vars. But the “bowing” on the biplot is a common problem when you have more than 1 env. gradient working in the data that are working at cross-purposes. Which you described in text, and which you get out of the regression (or classification) tree (which I did get backwards – it’s about the predictee, not the predictors, but not both). So my suggestion would be to stick with the CART analysis. If you must do a GLM, you should only work with uncorrelated BioClim vars. You’ll just have to choose the set a priori and defend it. Best, Aaron
<<<<<<< HEAD
Page 65:2016-09-12. variable importance
Page 66: 2016-09-13. climate cascade meeting
- Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
Multiple stressors ms:
- my hands, need to edit and send out by wednesday/thursday
Range limits ms: SHC's hands
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- update: Curtis can no longer work+ write on project
There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing and then get everything signed.
Go over thesis layout next time
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agreee
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agreee
Page 68: 2016-09-14. SICB meeting
Venue: Hilton New Orleans Riverside
Address: Two Poydras Street, New Orleans, LA 70130, UNITED STATES
Closest airport: Louis Armstrong New Orleans Airport.
27 minutes away from hilton but there is discounted round trip airport trans: $40/person
Budget:
- $40 transportation (put 32 in budget)
- $388 flight
- $580 + taxes and fees hotel
- $ 99 registration to SICB
Page 68: 2016-09-19; 2016-09-20. Climate cascade meeting
Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
Multiple stressors ms:
- sent to SHC 2016-09-16
Range limits ms: SHC's hands
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: ETA end of the week (5/6 done)
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Go over thesis layout next time
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Abstract? I have one written up for NSF post doc fellowship
Page 69: 2016-09-21. qpcr redos for 18s rRNA
Table of colonies with unstable HSG as determined by linear regression (18s ~ Temp).
| colony | Df | SS | MS | F | p_value | |
|---|---|---|---|---|---|---|
| 5 | ALA1 | 1 | 2123420.91 | 2123420.91 | 8.054925 | 0.0218751 |
| 9 | Avon19-1 | 1 | 85577.02 | 85577.02 | 5.659013 | 0.0446244 |
| 15 | CJ2 | 1 | 860194.07 | 860194.07 | 26.944017 | 0.0008317 |
| 55 | GF34-1 | 1 | 9742336.46 | 9742336.46 | 45.449574 | 0.0001463 |
| 85 | LPR4 | 1 | 2802821.86 | 2802821.86 | 14.940584 | 0.0047729 |
others: Yates3, Duke8
Page 70: 2016-09-26. selecting poplar clones
Overall goal: Make a map highlighting climate gradient and plotting potential sites to select clones from. The magnitiude of the points will relate to the GSL.
General workflow
- Grab climate data and plot all sites
- Link previous dataset to a another dataset that has empirical GSL from either IH or Burlington.
- Make map
Climate data

Looks like PC1 (~55%) represents preicipitation to temperature seasonality axis and PC2 (19%) represents precipitation to overall temperature axis.
All possible sites

Subsetted sites
Looks like IH has both BF and BS data but Burlington doesn't

range of GSLs: 2.016667-4.833333 months
Table for previous fig
| PopCode | GSL | BS | BF | months |
|---|---|---|---|---|
| CBI | 84.20000 | 200.9000 | 116.7000 | 2.806667 |
| CLK | 62.00000 | 183.8889 | 121.8889 | 2.066667 |
| CPL | 67.57143 | 190.3571 | 122.7857 | 2.252381 |
| CYP | 61.85714 | 184.8571 | 123.0000 | 2.061905 |
| FIS | 77.80000 | 194.0000 | 116.2000 | 2.593333 |
| FNO | 70.11111 | 181.0000 | 110.8889 | 2.337037 |
| GAM | 64.76000 | 186.6400 | 121.8800 | 2.158667 |
| HWK | 68.80000 | 189.8500 | 121.0500 | 2.293333 |
| KAP | 68.75000 | 193.3125 | 124.5625 | 2.291667 |
| KEN | 145.00000 | 256.6000 | 111.6000 | 4.833333 |
| LLC | 136.95000 | 248.1500 | 111.2000 | 4.565000 |
| LON | 91.72727 | 208.7273 | 117.0000 | 3.057576 |
| MBK | 61.77778 | 182.3333 | 120.5556 | 2.059259 |
| NBY | 91.23529 | 210.4706 | 119.2353 | 3.041177 |
| NEG | 76.88636 | 195.6591 | 118.7727 | 2.562879 |
| OUT | 69.40000 | 188.1000 | 118.7000 | 2.313333 |
| RAD | 63.77778 | 181.7037 | 117.9259 | 2.125926 |
| SKN | 63.38462 | 184.8462 | 121.4615 | 2.112821 |
| TBY | 137.60000 | 250.1333 | 112.5333 | 4.586667 |
| TUR | 63.40000 | 186.5000 | 123.1000 | 2.113333 |
| UMI | 61.00000 | 182.0000 | 121.0000 | 2.033333 |
| WLK | 60.50000 | 175.0000 | 114.5000 | 2.016667 |
Page 71: 2016-09-26 and 2016-09-27. Climate cascade meeting
Project updates:
Gene expression project: on hold; focusing on 2 manuscripts (multiple stressors and range limits ms)
- Present some analyses
- Present some analyses
Multiple stressors ms:
- Working on SHC edits
- Working on SHC edits
Range limits ms: SHC lab meeting to go over Thursday September 29th
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: ETA end of the week (5/6 done); database searching
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
Page 72: 2016-09-27. evolution of hsp gxp data analysis
Exploring different approaches
- PCA decomp bioclim variables I think a priori are important and using that in regression vs. just bio5(Tmax)
- And then building a global model with predictors I think are important (a priori) vs constructing a fully complex model.
Exploring hsp gxp parameters from boltzmann fits
Table of correlation between params
| FC_hsc701468max | FC_hsc701468slope | FC_hsc701468Tm | FC_hsp40541max | FC_hsp40541slope | FC_hsp40541Tm | FC_Hsp83279max | FC_Hsp83279slope | FC_Hsp83279Tm | |
|---|---|---|---|---|---|---|---|---|---|
| FC_hsc701468max | 1.000 | 0.569 | 0.642 | 0.398 | 0.104 | 0.076 | 0.029 | -0.122 | -0.207 |
| FC_hsc701468slope | 0.569 | 1.000 | 0.640 | 0.340 | 0.189 | 0.174 | -0.154 | -0.079 | -0.297 |
| FC_hsc701468Tm | 0.642 | 0.640 | 1.000 | 0.429 | 0.207 | 0.401 | -0.130 | -0.124 | -0.264 |
| FC_hsp40541max | 0.398 | 0.340 | 0.429 | 1.000 | 0.602 | 0.624 | 0.030 | 0.117 | -0.082 |
| FC_hsp40541slope | 0.104 | 0.189 | 0.207 | 0.602 | 1.000 | 0.651 | 0.037 | 0.122 | -0.129 |
| FC_hsp40541Tm | 0.076 | 0.174 | 0.401 | 0.624 | 0.651 | 1.000 | -0.247 | -0.075 | -0.215 |
| FC_Hsp83279max | 0.029 | -0.154 | -0.130 | 0.030 | 0.037 | -0.247 | 1.000 | 0.756 | 0.669 |
| FC_Hsp83279slope | -0.122 | -0.079 | -0.124 | 0.117 | 0.122 | -0.075 | 0.756 | 1.000 | 0.864 |
| FC_Hsp83279Tm | -0.207 | -0.297 | -0.264 | -0.082 | -0.129 | -0.215 | 0.669 | 0.864 | 1.000 |
It doesn't have basal gxp; including basal and then doing a pca:
Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7 Comp.8 Comp.9 Comp.10Standard deviation 2.0502371 1.4176264 1.2325728 1.1396205 0.84813044 0.74749858 0.68915615 0.60025005 0.4704591 0.36055794Proportion of Variance 0.3612359 0.1727056 0.1305593 0.1116100 0.06181701 0.04801793 0.04081483 0.03096329 0.0190207 0.01117205Cumulative Proportion 0.3612359 0.5339415 0.6645008 0.7761108 0.83792784 0.88594577 0.92676060 0.95772389 0.9767446 0.98791664 Comp.11 Comp.12Standard deviation 0.294315885 0.232345681Proportion of Variance 0.007444064 0.004639294Cumulative Proportion 0.995360706 1.000000000| Comp.1 | Comp.2 | Comp.3 | Comp.4 | |
|---|---|---|---|---|
| hsc70 | 0.366 | 0.117 | -0.041 | -0.400 |
| hsp83 | 0.271 | 0.019 | -0.238 | -0.414 |
| hsp40 | 0.141 | 0.309 | -0.279 | -0.433 |
| FC_hsc701468max | 0.284 | -0.006 | 0.529 | -0.184 |
| FC_hsc701468slope | 0.313 | 0.112 | 0.318 | -0.110 |
| FC_hsc701468Tm | 0.300 | -0.063 | 0.495 | 0.234 |
| FC_hsp40541max | 0.210 | -0.502 | 0.039 | -0.185 |
| FC_hsp40541slope | 0.153 | -0.521 | -0.264 | -0.048 |
| FC_hsp40541Tm | 0.232 | -0.493 | -0.175 | 0.173 |
| FC_Hsp83279max | -0.321 | -0.174 | 0.305 | -0.324 |
| FC_Hsp83279slope | -0.355 | -0.249 | 0.168 | -0.366 |
| FC_Hsp83279Tm | -0.392 | -0.124 | 0.124 | -0.276 |
some stats with pcas of hsp gxp params to see how much it explains CTmax
summary(lm(h$KO_temp_worker~paramspc$scores[,1]+paramspc$scores[,2]+paramspc$scores[,3]+paramspc$scores[,4]))Call:lm(formula = h$KO_temp_worker ~ paramspc$scores[, 1] + paramspc$scores[, 2] + paramspc$scores[, 3] + paramspc$scores[, 4])Residuals: Min 1Q Median 3Q Max -0.90448 -0.46768 0.02901 0.40598 1.08398 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 41.61667 0.10803 385.219 < 2e-16 ***paramspc$scores[, 1] 0.15861 0.05269 3.010 0.00548 ** paramspc$scores[, 2] -0.04312 0.07621 -0.566 0.57600 paramspc$scores[, 3] 0.40672 0.08765 4.640 7.41e-05 ***paramspc$scores[, 4] 0.05244 0.09480 0.553 0.58451 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.6206 on 28 degrees of freedomMultiple R-squared: 0.5272, Adjusted R-squared: 0.4596 F-statistic: 7.805 on 4 and 28 DF, p-value: 0.0002339Mistake: I didnt control for housekeeping gene in basal gxp. redo
h<-read.csv("20160927_total_dataset_curated.csv")basalxp<-h[,4:6]-h[,3]paramspc<-princomp(scale(cbind(basalxp,h[,7:15])))summary(paramspc)Importance of components: Comp.1 Comp.2 Comp.3 Comp.4 Comp.5Standard deviation 1.8681865 1.5167796 1.3456615 1.1162675 0.86282525Proportion of Variance 0.3002255 0.1979028 0.1557682 0.1071874 0.06404021Cumulative Proportion 0.3002255 0.4981283 0.6538964 0.7610838 0.82512400 Comp.6 Comp.7 Comp.8 Comp.9Standard deviation 0.82407918 0.71977398 0.55118644 0.45902754Proportion of Variance 0.05841776 0.04456556 0.02613389 0.01812527Cumulative Proportion 0.88354177 0.92810732 0.95424121 0.97236649 Comp.10 Comp.11 Comp.12Standard deviation 0.39039533 0.304603562 0.275767584Proportion of Variance 0.01311041 0.007981362 0.006541743Cumulative Proportion 0.98547690 0.993458257 1.000000000knitr::kable(round(paramspc$loadings[,1:4],3))| Comp.1 | Comp.2 | Comp.3 | Comp.4 | |
|---|---|---|---|---|
| hsc70 | -0.338 | 0.071 | -0.410 | -0.299 |
| hsp83 | -0.275 | 0.237 | -0.295 | -0.234 |
| hsp40 | -0.098 | 0.057 | -0.476 | -0.391 |
| FC_hsc701468max | -0.316 | -0.358 | 0.172 | -0.246 |
| FC_hsc701468slope | -0.195 | -0.360 | 0.211 | -0.172 |
| FC_hsc701468Tm | -0.289 | -0.347 | 0.253 | -0.044 |
| FC_hsp40541max | -0.414 | 0.177 | 0.147 | 0.127 |
| FC_hsp40541slope | -0.310 | 0.265 | -0.087 | 0.461 |
| FC_hsp40541Tm | -0.348 | 0.304 | 0.081 | 0.313 |
| FC_Hsp83279max | -0.390 | 0.076 | 0.292 | -0.183 |
| FC_Hsp83279slope | 0.053 | 0.439 | 0.418 | -0.286 |
| FC_Hsp83279Tm | 0.193 | 0.406 | 0.290 | -0.410 |
Stats
summary(stepAIC(lm(h$KO_temp_worker~paramspc$scores[,1]+paramspc$scores[,2]+paramspc$scores[,3]+paramspc$scores[,4]),direction="both"))Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 41.62969 0.12246 339.931 < 2e-16 ***paramspc$scores[, 1] -0.17806 0.06555 -2.716 0.01119 * paramspc$scores[, 2] -0.23931 0.08074 -2.964 0.00614 ** paramspc$scores[, 3] 0.15733 0.09101 1.729 0.09486 . ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.6928 on 28 degrees of freedomMultiple R-squared: 0.4062, Adjusted R-squared: 0.3425 F-statistic: 6.384 on 3 and 28 DF, p-value: 0.00196Page 73: 2016-09-28. building ultrametric trees
I need to build ultrametric trees to do phylogenetic analyses. They need to be ultrametric to meet the assumptions of Homoscedasticity. I'll be using BEAST 2.3.1. And I'll build 2 types; 1 with BL differences across whole phylogeny and another with species as polytomies.
I created a new folder in /Data/Phylogenetics/20160928_beast
It has 2 newick files: 20160927phylogeny_aphaeno_BL_species.newick and 20160927phylogeny_aphaeno_BL.newick
- 20160927phylogeny_aphaeno_BL.newick has BL for each colony, and I previously added CJ10, LPR4, and Bing in there; so I have to take them out because there is no sequence data for those samples. New file: 20160927phylogeny_aphaeno_BL_none.newick
- 20160927phylogeny_aphaeno_BL.newick has BL for each colony, and I previously added CJ10, LPR4, and Bing in there; so I have to take them out because there is no sequence data for those samples. New file: 20160927phylogeny_aphaeno_BL_none.newick
It also has this fasta file that was previously parsed: 20160516_Andrew_SNP_sequences.fas
In downstream analyses, I got rid of novomessor which I'll do for this fasta too. New file: 20160516_Andrew_SNP_sequences_nonov.fas
I'll use BEAST on cipres, but I'll need to set up Beauti which sets up the input for BEAST.
The two trees:
Pop level

species

Yes, so you can't put tree information into a beast analysis I dont think. Anyway, here are the settings:
NOte Need to convert fasta into nexus file in order for beauti to read as nucleotide, otherwise it'll read it as amino acids**
- 1 partition,(1 SNP matrix)
- tip dates specified as year and before the present
- gammer site model, GTR+gamma
- Relaxed clock log normal
- priors: ingropu = aphaenogaster, outgroup= veromessor; ucldMean set as mean = 50, sigma = 5 based on fossil data
- MCMC chain length = 100,000,000
Cannot get it to work. YULE model best for species. But I have pop and species.
Page 74: 2016-09-28. phylogenetic regressions (PGLS) and anovas
Did PGLS in 3 ways:
- untransformed BL
- transformed for all tips
- forced polytomies for species
1. untransformed BL

PGLS 1. untransformed BL
pgmod<-gls(KO_temp_worker~ bio5*habitat_v2, correlation = corBrownian(phy = aph_onlytree),data = aph_phylo, method = "ML")summary(pgmod)Generalized least squares fit by maximum likelihood Model: KO_temp_worker ~ bio5 * habitat_v2 Data: aph_phylo AIC BIC logLik 289.458 302.4838 -139.729Correlation Structure: corBrownian Formula: ~1 Parameter estimate(s):numeric(0)Coefficients: Value Std.Error t-value p-value(Intercept) 39.60864 3.41576 11.595866 0.0000bio5 0.00663 0.00968 0.685406 0.4947habitat_v2flat woods 9.03418 50.92693 0.177395 0.8596bio5:habitat_v2flat woods -0.02718 0.15809 -0.171921 0.8639 Correlation: (Intr) bio5 hbt_2wbio5 -0.870 habitat_v2flat woods -0.017 0.041 bio5:habitat_v2flat woods 0.016 -0.042 -1.000Standardized residuals: Min Q1 Med Q3 Max -1.52993175 -0.23380594 -0.04718187 0.06754746 0.45099889 Residual standard error: 2.995514 Degrees of freedom: 100 total; 96 residualPhyl ANOVA 1. untransformed BL
phlaov<-phylANOVA(aph_onlytree,aph_phylo$habitat_v2,aph_phylo$KO_temp_worker,p.adj="hochberg")phlaov$F[1] 49.0392$Pf[1] 0.135$T deciduous forest flat woodsdeciduous forest 0.000000 -7.002799flat woods 7.002799 0.000000$method[1] "hochberg"$Pt deciduous forest flat woodsdeciduous forest 1.000 0.135flat woods 0.135 1.0002. transformed for all tips

PGLS 2. transformed for all tips
ult.tree1<-compute.brlen(aph_onlytree)plot(ult.tree1,cex=.5)aph_phylo1<-ant_dat_clim[match(ult.tree1$tip.label,ant_dat_clim$colony.id2),]pgmod1<-gls(KO_temp_worker~ bio5*habitat_v2, correlation = corBrownian(phy = ult.tree1),data = aph_phylo1, method = "ML")summary(pgmod1)Generalized least squares fit by maximum likelihood Model: KO_temp_worker ~ bio5 * habitat_v2 Data: aph_phylo1 AIC BIC logLik 335.9159 348.9418 -162.958Correlation Structure: corBrownian Formula: ~1 Parameter estimate(s):numeric(0)Coefficients: Value Std.Error t-value p-value(Intercept) 39.94706 4.77385 8.367890 0.0000bio5 0.00486 0.01258 0.386220 0.7002habitat_v2flat woods 14.08505 51.06703 0.275815 0.7833bio5:habitat_v2flat woods -0.04342 0.15883 -0.273386 0.7851 Correlation: (Intr) bio5 hbt_2wbio5 -0.806 habitat_v2flat woods -0.008 0.025 bio5:habitat_v2flat woods 0.008 -0.025 -1.000Standardized residuals: Min Q1 Med Q3 Max -0.836519826 -0.092391337 0.004278385 0.080275482 0.347423662 Residual standard error: 5.332277 Degrees of freedom: 100 total; 96 residuaPHYLO ANOVA 2. transformed for all tips
phlaov2<-phylANOVA(ult.tree1,aph_phylo$habitat_v2,aph_phylo$KO_temp_worker,p.adj="hochberg")phlaov2$F[1] 49.0392$Pf[1] 0.234$T deciduous forest flat woodsdeciduous forest 0.000000 -7.002799flat woods 7.002799 0.000000$method[1] "hochberg"$Pt deciduous forest flat woodsdeciduous forest 1.000 0.234flat woods 0.234 1.0003. forced polytomies for species

PGLS 3. forced polytomies for species
plot(aph_onlytree)nodelabels(cex=.5)ant_tree_root1<-collapse.to.star(ant_tree_root,192) # florant_tree_root2<-collapse.to.star(ant_tree_root1,184) #ashant_tree_root3<-collapse.to.star(ant_tree_root2,158) #piceaant_tree_root4<-collapse.to.star(ant_tree_root3,131)# rudisant_tree_root5<-collapse.to.star(ant_tree_root4,119) # miamianaant_tree_root6<-collapse.to.star(ant_tree_root5,116) #lamellidensant_tree_root7<-collapse.to.star(ant_tree_root6,104) # fulvaant_tree_root8<-collapse.to.star(ant_tree_root7,103) # tenn#ant_tree_root9<-collapse.to.star(ant_tree_root8) # outgroupplot(ant_tree_root8)ult2.tree<-compute.brlen(ant_tree_root8)plot(ult2.tree)aph_phylo2<-ant_dat_clim[match(ult2.tree$tip.label,ant_dat_clim$colony.id2),]pgmod2<-gls(KO_temp_worker~bio5*habitat_v2, correlation = corBrownian(phy = ult2.tree),data = aph_phylo2, method = "ML")summary(pgmod2)Generalized least squares fit by maximum likelihood Model: KO_temp_worker ~ bio5 * habitat_v2 Data: aph_phylo2 AIC BIC logLik 255.776 268.8019 -122.888Correlation Structure: corBrownian Formula: ~1 Parameter estimate(s):numeric(0)Coefficients: Value Std.Error t-value p-value(Intercept) 37.82400 2.043758 18.507082 0.0000bio5 0.01175 0.005942 1.978037 0.0508habitat_v2flat woods 22.58447 12.917075 1.748420 0.0836bio5:habitat_v2flat woods -0.06971 0.039823 -1.750585 0.0832 Correlation: (Intr) bio5 hbt_2wbio5 -0.881 habitat_v2flat woods -0.132 0.148 bio5:habitat_v2flat woods 0.132 -0.149 -0.999Standardized residuals: Min Q1 Med Q3 Max -2.24865470 -0.26276358 0.05811258 0.26246427 0.99070867 Residual standard error: 1.836591 Degrees of freedom: 100 total; 96 residualPHYLO ANOVA 3. forced polytomies for species
aph_phylo<-ant_dat_clim[match(ult2.tree$tip.label,ant_dat_clim$colony.id2),]aph_phylo$habitat_v2<-droplevels(aph_phylo$habitat_v2)phlaov3<-phylANOVA(ult2.tree,aph_phylo$habitat_v2,aph_phylo$KO_temp_worker,p.adj="hochberg")phlaov3$F[1] 49.0392$Pf[1] 0.183$T deciduous forest flat woodsdeciduous forest 0.000000 -7.002799flat woods 7.002799 0.000000$method[1] "hochberg"$Pt deciduous forest flat woodsdeciduous forest 1.000 0.183flat woods 0.183 1.000Intepretting a phylogenetic ANOVA here
The way the phylogenetic ANOVA (sensu Garland et al. 1993; Syst. Biol.) works is by first computing a standard ANOVA, and then comparing the observed F to a distribution obtained by simulating on the tree under a scenario of no effect of x on y. This "accounts for" the tree in the sense that it attempts to account for the possibility that species may have similar y conditioned on x because x influences y; or because they share common history and are thus similar by virtue of this history (and not at all due to x) It is not particularly surprising that your P-value was lower in the phylogenetic ANOVA than in your regular ANOVA. In general, the effect of the phylogenetic ANOVA on P depends on the distribution of the factor, x. If x is clumped on the tree, than the P-value of a phylogenetic ANOVA will tend to be higher than a regular ANOVA. By contrast, if x is overdispersed phylogenetically, the P-value of the phylogenetic ANOVA will tend to be lower than the regular ANOVA. I hope this is of some help. - Liam -- Liam J. Revell University of Massachusetts Boston web: http://faculty.umb.edu/liam.revell/ email: ***@umb.edu blog: http://phytools.blogspot.com
2016-09-28. SHC suggestion: ancestral trait reconsturction -> regressions/anovas
2016-09-29. PIC
Dataset
| Species | CTmax | Tmax | Habitat | |
|---|---|---|---|---|
| 1 | ashmeadi | 42.80833 | 324.0000 | FW |
| 2 | floridana | 42.76852 | 323.7778 | FW |
| 6 | picea | 40.50096 | 262.9615 | DF |
| 7 | rudis | 41.33808 | 300.3214 | DF |
| 5 | miamiana | 40.95128 | 329.3846 | DF |
| 4 | lamellidens | 42.09375 | 318.2500 | DF |
| 3 | fulva | 41.01222 | 310.5556 | DF |
| 8 | tennesseensis | 40.75000 | 311.0000 | DF |
NOdes of phylogeny

Independent contrast estimates for CTMAX
| Node | cctmax |
|---|---|
| 1001 | -0.7004417 |
| 1002 | -0.5834076 |
| 1003 | -1.5296702 |
| 1004 | 0.8678094 |
| 1005 | 0.2051669 |
| 92 | 2.0095026 |
| 1006 | -0.0679396 |
Better fig with contrsts of CTmax v Tmax with points labeled by nodes

Page 75: 2016-10-03 and 2016-10-04.Climate cascade meeting
Project updates:
Gene expression project:
Go over analyses:
- Phylo anova, PGLS, ancestral trait reconstruction
- GXP: basal expression, PGLS with CTmax and gxp parameters
Go over figure layout for ms
Left to do: QC and analyze hsp83 and hsp40
Multiple stressors ms:
- Ask about SHC comments on confusion of mismatch membrane stability
- Ask about SHC comments on confusion of mismatch membrane stability
Range limits ms: SHC lab gave verbal edits:
- focus on 1 end of thermal niche breadth(although it is nice to mention it because CTmin decreases across lat)--CTmin.
- Dicussion needs to talk about cold adaptation; why trade-offs?
- Walk through results better
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: no clue what status is
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Talk title: Northern range limits of a common forest ant is associated with trade-offs in cold physiology
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 76: 2016-10-03. Membrane stability
Trying to get how membrane stability is altered among different stressors. Two things can change/alter membrane fluidity; glycero-phospholipid head groups (phosphotidylethanolamine,PE; phosphotidylcholine PC) and lipid saturation(saturated vs unsaturated). In warmer environments, higher PC and lipid saturation confer homeostasis. Cooler environments = PE and unsaturated lipids. Membrane fluidity for desiccation resistance usually covaries with cold acclimation/adaptation.
PC bind 10-12 water molecules and PE binds 7-8. PE binds less water and it should be enriched under desiccation stress.

Going through some of the literature and what they found.
Hayward et al. 2014 has a nice explanation

In A, those that have greater unsaturated fatty acids, are more cold tolerant (operative body temperature to be exact). More fatty acid content negatively correlated with DPH anisotropy at 20 C (something that distorts light). DPH related to membrane rigidity and fluidity; high values = reduced constraint on intra-membrane molecular mobility. So High unsaturated fatty acid index is related to reduced membrane rigidity or high membrane fluidity (lower values of DPH anisotropy)
Cossins and Prosser 1978 PNAS shows:

High membraine fluidity (polarization) is higher in more cold acclimated fish. and...

High membrane fluidity is related to higher saturated:unsaturated. I think this makes sense, high unsat FA makes the polarization smaller.
Cooper et al. 2014; Functional ecology finds that acclimation ifluences PE/PC ratios

Summary table of directions of effects for stressors on membrane fluidity
| Stress.type | Heat | Cold | Desiccation | pH |
|---|---|---|---|---|
| membrane.fluidity | decrease | increase | increase | |
| membrane.rigidity | increase | decrease | decrease | |
| PC | increase | decrease | decrease | |
| PE | decrease | increase | increase | |
| PE.PC.ratio | decrease | increase | increase | |
| saturated.FA | increase | decrease | decrease | |
| unsaturated.FA | decrease | increase | increase | |
| saturated.unsaturated.ratio | increase | decrease | decrease |
refs:
Hazel, J. R., and E. Eugene Williams. 1990. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Progress in Lipid Research 29:167–227.
Hazel, J. R., S. J. McKinley, and M. F. Gerrits. 1998. Thermal acclimation of phase behavior in plasma membrane lipids of rainbow trout hepatocytes. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology 275:R861–R869.
Cooper, B. S., L. A. Hammad, and K. L. Montooth. 2014. Thermal adaptation of cellular membranes in natural populations of Drosophila melanogaster. Functional Ecology 28:886–894.
Cossins, A. R., and C. L. Prosser. 1978. Evolutionary adaptation of membranes to temperature. Proceedings of the National Academy of Sciences of the United States of America 75:2040–2043.
Hayward, S. A. L., B. Manso, and A. R. Cossins. 2014. Molecular basis of chill resistance adaptations in poikilothermic animals. The Journal of Experimental Biology 217:6–15.
Holmstrup, M., K. Hedlund, and H. Boriss. 2002. Drought acclimation and lipid composition in Folsomia candida: implications for cold shock, heat shock and acute desiccation stress. Journal of Insect Physiology 48:961–970.
Page 77: 2016-10-04 Lab Safety Officer (LSO) meeting.
Department of Risk Management and Safety- Francis Churchill mainly speaking
Agenda:
1. News and updates
staff changes- new lab safety coordinator
lab fires at uvm
Chemistry- no blame; removing syringe that had fire . no evac, not a big fire
Votey building - small fire; no damage no hurt; alcohol near a burner--fire
- faculty said not to leave in 1 class; that is bad. You should leave if fire alarm goes off.
- faculty said not to leave in 1 class; that is bad. You should leave if fire alarm goes off.
Explosion at U Hawaii
- post doc in lab; working with pressure vessel (creating fuel for bacteria to make biofilms and biofuels); mixing hydrogen and oxygen and some carbon dioxide. Did over and over, and had minor issues; but in march it blew up. Took her arm off. Lab had good safety; but regulatory agents don't know how stuff get mixed; we all need to get better at hazard assessment. Fined [Math Processing Error]750,000 building damage. Brought up issue of coverage of isurance for post doc researchers
- Violations: Failed t provide a safe workplace; failed to ensure employees to follow proper procedures. Chemical Hygiene plan did not include SOPs for relevant safety.
- post doc in lab; working with pressure vessel (creating fuel for bacteria to make biofilms and biofuels); mixing hydrogen and oxygen and some carbon dioxide. Did over and over, and had minor issues; but in march it blew up. Took her arm off. Lab had good safety; but regulatory agents don't know how stuff get mixed; we all need to get better at hazard assessment. Fined [Math Processing Error]750,000 building damage. Brought up issue of coverage of isurance for post doc researchers
Fine at Oregon
- $275,000 by EPA for mismanagement of chemicals; did not get rid of their chemicals; no labeled; every bottle out there should be labeled.
- $275,000 by EPA for mismanagement of chemicals; did not get rid of their chemicals; no labeled; every bottle out there should be labeled.
We're going to be inspected by the US EPA and the state department(DEC)
risk control governance: 22% of safety trainings are not being completed; high for lab supervisors!!!! Lab safety notebooks need to be updated.
2. uvm police services
Office Sue Roberts: Work place violence. Active shooters? Training to safeguard to active shooters. How to respond?
Showing a video: Run, Hide, Fight. Know how to exit your building(how to get in or out). First responders don't tend to the injured; secondary responders will.
Systems in place:
- own police agency on campus with master keys and card access; allowing quick response times.
- CAT Alerts.
- Emergency blue lights- direct connect to UVM police.
Violence in the workplace
- Detect early, to get resoureces to person with alarming behavior.
- 2 teams on campus meet weekly and monthly; safety response team (discuss faculty and staff on campus) and care team (focus on needs of students). There is an anonymous care form (please give tons of info).
Phone systems
- for lan line; 911 goes to uvm police and they know where you are, send office to location
- for cell phones it goes to 911 call center in williston or it could go to brattoboro. Pay attention to where you are because phones don't give you pinpoint accuracy. Know street address.
- Put UVM police into contact list: 802-656-3473
3. summary of audits
There are top 10 audit deficiencies: FILL OUT DATES; use yellow waste label
- safety training incomplete
- chemical waste is older than 6 months (we need a sticker and they need to collect the waste)
- mislabelling in chemical waste containers (completely fill out tags!!)
- reports of hazard assessments are not available
- lab online inventory (HCOC) has not been updated wtihin 6 months
- chemical containers not fully labeled (Waste and non waste need labels)
- research samples not albeled properly: sample ID, hazards, date material made
- info on emergency contact door is not current
- lab monthly inspection not done
- eyewash flush log not visible and current.
Creating corrective actions: Stuff for you to fix.:
4. lab safety basics
UVM lab safety; monthly self inspection: Policy, all labs must do monthly inspection. Document on checklist.
If you don't have one, they are distributed out to departments.
- Defrost freezers. check website so that our freezer is not ruined.
- Label samples
- annual refresher training ( everybody complete it? )
- can write descrepancies.
Labels: You need manufacturer's label and don't need anythingelse, just sign and date it.
Safety Audits at UVM: LabCliq. LSO can do corrective actions but the PI has to use Labcliq to verify online. Then PI gets email.
What trainings do you need? HERE
- Take all things that are applicable to your laboratory!!
- Green section 6 classes+ Annual refresher training. 4 online safety trainings and 2 classroom safety trainings.
- Red section Fire safety training.
Lab safety notebook webpage HERE
5. CITI training opportunity
6. Q & A
- Fraudulent calls: Target international faculty and staff referring to immigration status, healthcare, taxes. If you get calls, notify police services to set up trap on that phone. CHeck for scams on UVM police website
Page 78: 2016-10-05. Hsp gxp function valued trait fig
Boltzmann function and fit to dataset
Boltz<-function(data=x){ B<-nls(gxp ~ (1+(max-1)/(1+exp((Tm-T)/a))),data=data, start=list(max=80,Tm=35,a=1.05), trace=TRUE,control=nls.control(warnOnly = TRUE, tol = 1e-05, maxiter=1000))#summary(B) return(summary(B)$parameters)}T<-c(25,28,30,31.5,33,35,36.5,38.5,40,41)gxp<-c(1.050927323,1.795269722,2.394945916,2.025719648,5.995719441,12.75328258,35.0828896,44.80226791,63.64704198,67.607218)FB1<-as.data.frame(cbind(T,gxp));FB1Boltz(FB1)knitr::kable(Boltz(FB1))function that estimates values based on Boltzmann parameters
fud<-function(T=seq(25,70,.1),Tm=40,slope=1.8,max=50){ y<-1+ (max-1)/(1+exp(((Tm-T)/slope))) return(y) }parameter fits
| Estimate | Std. Error | t value | p value | |
|---|---|---|---|---|
| max | 76.179606 | 8.0617514 | 9.449511 | 0.0000310 |
| Tm | 37.432787 | 0.5585165 | 67.021804 | 0.0000000 |
| a | 1.765851 | 0.3248254 | 5.436310 | 0.0009701 |
With units and real data
plot(seq(0,70,.1),fud(T=seq(0,70,.1)),col="blue",type="n",ylim=c(0,80),las=1,xlab="",ylab="",xlim=c(25,42))mtext("Fold Induction", side=2, line=2.5, cex=2)mtext("Temperature", side=1, line=2.7, cex=2)lines(seq(25,70,.1),fud(Tm=37.4,slope=1.76,max=76),lwd=6)points(FB1$T,FB1$gxp,pch=19,col="blue",cex=3)lines(c(37.4,37.4),c(-10,39),lwd=5,lty="dotdash",col="purple")abline(h=73,lty="dotdash",col="red",lwd=5)arrows(35,20,38,50,code=2,lwd=2,)text(c(39,30,36),c(20,76,50),c("Tm","Max","Slope"),font=2,cex=2)
No units or real data
plot(seq(0,70,.1),fud(T=seq(0,70,.1)),col="blue",type="n",ylim=c(0,80),las=1,xlab="",ylab="",xlim=c(25,42),axes=FALSE)mtext("Fold Induction", side=2, line=2.5, cex=2)mtext("Temperature", side=1, line=2.7, cex=2)lines(seq(25,70,.1),fud(Tm=37.4,slope=1.76,max=76),lwd=6)#points(FB1$T,FB1$gxp,pch=19,col="blue",cex=3)lines(c(37.4,37.4),c(-10,39),lwd=5,lty="dotdash",col="purple")abline(h=73,lty="dotdash",col="red",lwd=5)arrows(35,20,38,50,code=2,lwd=2,)text(c(39,30,36),c(20,76,50),c("Tm","Max","Slope"),font=2,cex=2)box()
Page 79: 2016-10-06. SHC lab meeting: NSF post doc app
Lab safety stuff:
- Do trainings online
- Check waste and dispose it, ethidium bromide gels
- Do monthly inspections
Newar works on Fridays; works up to 6 hours.
Notes:
- use performance curves or reaction norm instead of function-valued traits
- separate out terms, performance for fitness proxy and then reaction norm for physiology or any traits-phenology GxE = reaction norm; generate performance cruve--growth over season
- context depdnent expression of traits drive relative performance
- who cares about separating out photoperiod vs temp
- env can shape relationship between traits and performance in non-linera and unexpected ways or in ways that influence the process of adaptation, adaptive potential.
- look at many gxp traits-relating those to each other and to performance
- integrate all of these traits and overlay them on a complex environmental background
- stoichiometry: give ratios not just %
- expand on QG of gene expression
- selection may act in context-dependent manner
- be careful about constraints and trade-offs
- Think about training objective # 3; goal of grant? reword to make sure its a goal
- certain clones: does not tell you a whole lot. how should poplar be selected? Talk about general principals that you can lead to suggest to growers. What kind of outreach . prescribe based on environmental variables I am measuring.
- more info that is concrete on what the patterns are; feels adrift; not tied tightly between sections
- introduction- get rid of 2nd paragraph. maybe 1 sentence to previous paragraph
- reserach objectives: clarify traits; response function; add a little bit or shift; clarify parts
- get the realized GSL ; using existing rad seq data; predict performance as a function of temperature
Page 80: 2016-10-07. Prepping cliamte cascade meeting
Project updates:
Gene expression project:
- Go over analyses:
- Go over figure layout for ms
Multiple stressors ms:
- Ask about SHC comments on confusion of mismatch membrane stability
- Ask about SHC comments on confusion of mismatch membrane stability
Range limits ms: SHC lab gave verbal edits:
- focus on 1 end of thermal niche breadth(although it is nice to mention it because CTmin decreases across lat)--CTmin.
- Dicussion needs to talk about cold adaptation; why trade-offs?
- Walk through results better
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: no clue what status is
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Talk title: Northern range limits of a common forest ant is associated with trade-offs in cold physiology
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 81: 2016-10-11. ANCOVA models for testing interaction of hsp gxp parameter and habitat on CTmax
apply(b2[,3:11],2,function(x){summary(aov(b2$KO_temp_worker~b2$habitat_v2*x))})$FC_hsc70_1468_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 20.902 20.902 81.798 1.1e-11 ***x 1 0.375 0.375 1.467 0.232 b2$habitat_v2:x 1 0.374 0.374 1.462 0.233 Residuals 45 11.499 0.256 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsc70_1468_slope Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 20.902 20.902 84.903 6.33e-12 ***x 1 1.169 1.169 4.749 0.0346 * b2$habitat_v2:x 1 0.000 0.000 0.000 0.9999 Residuals 45 11.078 0.246 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsc70_1468_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 20.902 20.902 89.676 2.79e-12 ***x 1 1.125 1.125 4.828 0.0332 * b2$habitat_v2:x 1 0.633 0.633 2.718 0.1062 Residuals 45 10.489 0.233 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsp40_541_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 21.311 21.311 85.111 9.4e-12 ***x 1 0.360 0.360 1.440 0.2368 b2$habitat_v2:x 1 0.875 0.875 3.494 0.0684 . Residuals 43 10.767 0.250 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_hsp40_541_slope Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 21.311 21.311 81.495 1.75e-11 ***x 1 0.605 0.605 2.312 0.136 b2$habitat_v2:x 1 0.153 0.153 0.585 0.449 Residuals 43 11.245 0.262 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_hsp40_541_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 21.311 21.311 104.527 4.39e-13 ***x 1 1.642 1.642 8.052 0.00691 ** b2$habitat_v2:x 1 1.594 1.594 7.816 0.00771 ** Residuals 43 8.767 0.204 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_Hsp83_279_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 23.226 23.226 95.284 8.72e-13 ***x 1 0.063 0.063 0.260 0.612 b2$habitat_v2:x 1 0.330 0.330 1.355 0.250 Residuals 46 11.213 0.244 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 14 observations deleted due to missingness$FC_Hsp83_279_slope Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 23.226 23.226 95.648 8.22e-13 ***x 1 0.156 0.156 0.641 0.428 b2$habitat_v2:x 1 0.281 0.281 1.157 0.288 Residuals 46 11.170 0.243 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 14 observations deleted due to missingness$FC_Hsp83_279_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 23.226 23.226 95.177 8.88e-13 ***x 1 0.068 0.068 0.279 0.600 b2$habitat_v2:x 1 0.313 0.313 1.283 0.263 Residuals 46 11.225 0.244 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 14 observations deleted due to missingnessSummary table of each parameter and its interaction with habitat on CTmax:
| summary.table | max70 | slope70 | Tm70 | max40 | slope40 | Tm40 | max83 | slope83 | Tm83 |
|---|---|---|---|---|---|---|---|---|---|
| habitat | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| parameter | no | yes | yes | no | no | yes | no | no | no |
| habitat * parameter | no | no | no | no | no | yes | no | no | no |
Effect of habitat type on hsp gxp parameters
apply(b2[,3:11],2,function(x){summary(aov(x~b2$habitat_v2))})$FC_hsc70_1468_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 4819 4819 30.98 1.21e-06 ***Residuals 47 7312 156 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsc70_1468_slope Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 2.562 2.5621 12.99 0.000754 ***Residuals 47 9.270 0.1972 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsc70_1468_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 18.41 18.409 25.53 7.03e-06 ***Residuals 47 33.89 0.721 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 15 observations deleted due to missingness$FC_hsp40_541_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 110.7 110.69 5.018 0.0301 *Residuals 45 992.5 22.06 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_hsp40_541_slope Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 2.683 2.683 4.294 0.044 *Residuals 45 28.123 0.625 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_hsp40_541_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 39.38 39.38 14.2 0.000476 ***Residuals 45 124.81 2.77 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 17 observations deleted due to missingness$FC_Hsp83_279_max Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 149.4 149.43 5.649 0.0215 *Residuals 48 1269.8 26.45 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 14 observations deleted due to missingness$FC_Hsp83_279_slope Df Sum Sq Mean Sq F value Pr(>F)b2$habitat_v2 1 1.92 1.9227 2.345 0.132Residuals 48 39.35 0.8198 4 observations deleted due to missingness$FC_Hsp83_279_Tm Df Sum Sq Mean Sq F value Pr(>F) b2$habitat_v2 1 42.56 42.56 9.229 0.00385 **Residuals 48 221.37 4.61 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 14 observations deleted due to missingnessSummary table of effect of habitat on hsp gxp parameter
| param | habitat |
|---|---|
| max70 | yes |
| slope70 | yes |
| Tm70 | yes |
| max40 | yes |
| slope40 | yes |
| Tm40 | yes |
| max83 | yes |
| slope83 | no |
| Tm83 | yes |
Page 82: 2016-10-11. variance partitioning in CTmax of aphaeno
- Phylogenetic axes = first 9
- Ecology = MAT, TMax, and habitat type
#model constructionvar2<- varpart(Aph.dat$KO_temp_worker, ~ Axis.1 + Axis.2+ Axis.3+ Axis.4+Axis.5+Axis.6+Axis.7+Axis.8+Axis.9, ~bio1+bio5+habitat_v2,data=Aph.dat)$part$SS.Y[1] 121.5443$fract Df R.squared Adj.R.squared Testable[a+b] = X1 9 0.5199228 0.4719151 TRUE[b+c] = X2 3 0.4388392 0.4213030 TRUE[a+b+c] = X1+X2 12 0.5288496 0.4638634 TRUE$indfract Df R.squared Adj.R.squared Testable[a] = X1|X2 9 NA 0.042560390 TRUE[b] 0 NA 0.429354679 FALSE[c] = X2|X1 3 NA -0.008051705 TRUE[d] = Residuals NA NA 0.536136636 FALSEFigure with different components

Page 83: 2016-10-12. Testing effect of MAT on Hsp gxp and looking at correlations between phylogeny and climate.
> apply(mergy[,38:43],2,function(x){summary(lm(log10(x)~mergy$bio1))})$FC_83Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -0.69315 -0.17367 -0.02182 0.16945 0.66741 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.993238 0.115874 8.572 1.19e-11 ***mergy$bio1 -0.000497 0.001227 -0.405 0.687 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.2879 on 54 degrees of freedomMultiple R-squared: 0.003028, Adjusted R-squared: -0.01543 F-statistic: 0.164 on 1 and 54 DF, p-value: 0.6871$FC_70Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -0.63143 -0.12966 0.02354 0.18406 0.45652 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.571710 0.105899 14.842 <2e-16 ***mergy$bio1 0.000679 0.001122 0.605 0.547 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.2631 on 54 degrees of freedomMultiple R-squared: 0.006742, Adjusted R-squared: -0.01165 F-statistic: 0.3666 on 1 and 54 DF, p-value: 0.5474$FC_40Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -0.87164 -0.16033 0.05806 0.23030 0.71656 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.8929016 0.1372969 6.503 2.63e-08 ***mergy$bio1 0.0002741 0.0014540 0.188 0.851 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3411 on 54 degrees of freedomMultiple R-squared: 0.0006575, Adjusted R-squared: -0.01785 F-statistic: 0.03553 on 1 and 54 DF, p-value: 0.8512$B_83Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -0.86395 -0.31896 -0.04139 0.33454 0.76906 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) 0.203307 0.186138 1.092 0.280mergy$bio1 -0.002098 0.001971 -1.064 0.292Residual standard error: 0.4624 on 54 degrees of freedomMultiple R-squared: 0.02054, Adjusted R-squared: 0.002405 F-statistic: 1.133 on 1 and 54 DF, p-value: 0.292$B_70Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -0.9569 -0.3399 -0.0464 0.3489 0.8581 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) 0.199005 0.172676 1.152 0.254mergy$bio1 -0.002843 0.001829 -1.555 0.126Residual standard error: 0.429 on 54 degrees of freedomMultiple R-squared: 0.04284, Adjusted R-squared: 0.02512 F-statistic: 2.417 on 1 and 54 DF, p-value: 0.1259$B_40Call:lm(formula = log10(x) ~ mergy$bio1)Residuals: Min 1Q Median 3Q Max -1.68902 -0.28172 0.07947 0.31104 0.98014 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) 0.300482 0.221888 1.354 0.181mergy$bio1 -0.003086 0.002350 -1.313 0.195Residual standard error: 0.5512 on 54 degrees of freedomMultiple R-squared: 0.03096, Adjusted R-squared: 0.01301 F-statistic: 1.725 on 1 and 54 DF, p-value: 0.1946Summary: none are significant
Correlation between Mean Annual Temperature (MAT), Tmax, and 4 phylogenetic axes
| MAT | Tmax | Axis.1 | Axis.2 | Axis.3 | Axis.4 | |
|---|---|---|---|---|---|---|
| MAT | 1.000 | 0.910 | 0.857 | 0.197 | 0.182 | 0.132 |
| Tmax | 0.910 | 1.000 | 0.836 | 0.128 | 0.204 | 0.110 |
| Axis.1 | 0.857 | 0.836 | 1.000 | 0.002 | 0.000 | 0.008 |
| Axis.2 | 0.197 | 0.128 | 0.002 | 1.000 | 0.000 | -0.002 |
| Axis.3 | 0.182 | 0.204 | 0.000 | 0.000 | 1.000 | 0.000 |
| Axis.4 | 0.132 | 0.110 | 0.008 | -0.002 | 0.000 | 1.000 |
20161013 follow up: checking 18s HKG stability
If there is an effect of rearing temperature, Tmax, and/or heat shock treatment, phylo axes, then the HKG is not stable.
ct<-read.csv("../Data/20150810_raw_CT_values.csv")z<-inner_join(ct,mergy,by="Colony")z$qpcr_block<-as.factor(z$qpcr_block)#different 18s ct among treatments?#different 18s ctsummary(stepAIC(lm(log2(X18)~bio5*treatment+qpcr_block+Axis.1+Axis.2+Axis.3+Rearing_Temp,data=z2)),direction="forward")Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 3.176814 0.056303 56.424 < 2e-16 ***qpcr_block2 0.107059 0.017592 6.086 1.84e-08 ***qpcr_block3 0.163280 0.018586 8.785 2.83e-14 ***Axis.1 -0.136572 0.072299 -1.889 0.0616 . Axis.2 0.204421 0.112195 1.822 0.0712 . Axis.3 -0.278600 0.165081 -1.688 0.0944 . Rearing_Temp -0.003763 0.002393 -1.573 0.1187 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.07137 on 107 degrees of freedomMultiple R-squared: 0.5648, Adjusted R-squared: 0.5404 F-statistic: 23.15 on 6 and 107 DF, p-value: < 2.2e-1620161013 Taking out Axis1 because it covaries with bio5(Tmax)
apply(mergy[,38:43],2,function(x){summary(stepAIC(lm(log10(x)~mergy$bio5+mergy$Rearing_Temp+mergy$Axis.2+mergy$Axis.3)),direction="forward")})Start: AIC=-142.41log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.3 1 0.002884 4.1926 -144.37- mergy$Axis.2 1 0.008699 4.1984 -144.29- mergy$bio5 1 0.017061 4.2068 -144.18<none> 4.1897 -142.41- mergy$Rearing_Temp 1 0.257200 4.4469 -140.96Step: AIC=-144.37log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 Df Sum of Sq RSS AIC- mergy$Axis.2 1 0.009219 4.2018 -146.25- mergy$bio5 1 0.021070 4.2137 -146.08<none> 4.1926 -144.37- mergy$Rearing_Temp 1 0.254448 4.4471 -142.96Step: AIC=-146.25log10(x) ~ mergy$bio5 + mergy$Rearing_Temp Df Sum of Sq RSS AIC- mergy$bio5 1 0.01849 4.2203 -147.99<none> 4.2018 -146.25- mergy$Rearing_Temp 1 0.29906 4.5009 -144.26Step: AIC=-147.99log10(x) ~ mergy$Rearing_Temp Df Sum of Sq RSS AIC<none> 4.2203 -147.99- mergy$Rearing_Temp 1 0.30548 4.5258 -145.94Start: AIC=-151.28log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.3 1 0.006133 3.6020 -153.18- mergy$bio5 1 0.014353 3.6102 -153.05- mergy$Axis.2 1 0.125441 3.7213 -151.29<none> 3.5959 -151.28- mergy$Rearing_Temp 1 0.211236 3.8071 -149.97Step: AIC=-153.18log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 Df Sum of Sq RSS AIC- mergy$bio5 1 0.011172 3.6132 -155.00<none> 3.6020 -153.18- mergy$Axis.2 1 0.128482 3.7305 -153.15- mergy$Rearing_Temp 1 0.218797 3.8208 -151.76Step: AIC=-155log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2 Df Sum of Sq RSS AIC<none> 3.6132 -155.00- mergy$Axis.2 1 0.13788 3.7510 -154.83- mergy$Rearing_Temp 1 0.22616 3.8393 -153.48Start: AIC=-127.73log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.3 1 0.03867 5.4351 -129.32- mergy$bio5 1 0.10859 5.5051 -128.58<none> 5.3965 -127.73- mergy$Axis.2 1 0.42509 5.8216 -125.33- mergy$Rearing_Temp 1 0.64013 6.0366 -123.23Step: AIC=-129.32log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 Df Sum of Sq RSS AIC- mergy$bio5 1 0.14392 5.5791 -129.80<none> 5.4351 -129.32- mergy$Axis.2 1 0.41361 5.8488 -127.06- mergy$Rearing_Temp 1 0.67128 6.1064 -124.56Step: AIC=-129.8log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2 Df Sum of Sq RSS AIC<none> 5.5791 -129.80- mergy$Axis.2 1 0.47047 6.0495 -127.11- mergy$Rearing_Temp 1 0.63445 6.2135 -125.56Start: AIC=-88.85log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.2 1 0.02655 10.576 -90.709- mergy$bio5 1 0.27432 10.824 -89.365<none> 10.549 -88.854- mergy$Axis.3 1 0.47944 11.029 -88.277- mergy$Rearing_Temp 1 0.48666 11.036 -88.239Step: AIC=-90.71log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$bio5 1 0.29726 10.873 -91.101<none> 10.576 -90.709- mergy$Rearing_Temp 1 0.46041 11.036 -90.237- mergy$Axis.3 1 0.49173 11.068 -90.073Step: AIC=-91.1log10(x) ~ mergy$Rearing_Temp + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.3 1 0.36201 11.235 -91.201<none> 10.873 -91.101- mergy$Rearing_Temp 1 0.50260 11.376 -90.480Step: AIC=-91.2log10(x) ~ mergy$Rearing_Temp Df Sum of Sq RSS AIC<none> 11.235 -91.201- mergy$Rearing_Temp 1 0.56062 11.796 -90.377Start: AIC=-126.78log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.2 1 0.0042 5.4901 -128.735- mergy$Axis.3 1 0.0404 5.5262 -128.354- mergy$bio5 1 0.1532 5.6391 -127.182<none> 5.4859 -126.780- mergy$Rearing_Temp 1 4.5602 10.0461 -93.689Step: AIC=-128.74log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$Axis.3 1 0.0392 5.5292 -130.323- mergy$bio5 1 0.1609 5.6509 -129.060<none> 5.4901 -128.735- mergy$Rearing_Temp 1 4.8078 10.2978 -94.254Step: AIC=-130.32log10(x) ~ mergy$bio5 + mergy$Rearing_Temp Df Sum of Sq RSS AIC<none> 5.5292 -130.323- mergy$bio5 1 0.204 5.7332 -130.221- mergy$Rearing_Temp 1 4.770 10.2992 -96.246Start: AIC=-80.6log10(x) ~ mergy$bio5 + mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC- mergy$bio5 1 0.1822 12.346 -81.733<none> 12.164 -80.595- mergy$Rearing_Temp 1 0.7613 12.925 -79.074- mergy$Axis.2 1 1.1960 13.360 -77.156- mergy$Axis.3 1 3.4308 15.595 -68.185Step: AIC=-81.73log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3 Df Sum of Sq RSS AIC<none> 12.346 -81.733- mergy$Rearing_Temp 1 0.8276 13.174 -79.970- mergy$Axis.2 1 1.3181 13.664 -77.849- mergy$Axis.3 1 3.9458 16.292 -67.648$FC_83Call:lm(formula = log10(x) ~ mergy$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.6121 -0.1422 -0.0417 0.1399 0.7465 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.39083 0.27720 1.410 0.1641 mergy$Rearing_Temp 0.02473 0.01228 2.013 0.0489 *---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.2745 on 56 degrees of freedomMultiple R-squared: 0.0675, Adjusted R-squared: 0.05084 F-statistic: 4.053 on 1 and 56 DF, p-value: 0.0489$FC_70Call:lm(formula = log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2)Residuals: Min 1Q Median 3Q Max -0.67832 -0.16434 0.02663 0.17901 0.38810 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 2.12428 0.26812 7.923 1.16e-10 ***mergy$Rearing_Temp -0.02197 0.01184 -1.855 0.0689 . mergy$Axis.2 0.81467 0.56233 1.449 0.1531 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.2563 on 55 degrees of freedomMultiple R-squared: 0.07529, Adjusted R-squared: 0.04167 F-statistic: 2.239 on 2 and 55 DF, p-value: 0.1162$FC_40Call:lm(formula = log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2)Residuals: Min 1Q Median 3Q Max -0.80408 -0.10662 0.07152 0.25390 0.55421 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.08767 0.33317 0.263 0.7934 mergy$Rearing_Temp 0.03680 0.01471 2.501 0.0154 *mergy$Axis.2 1.50486 0.69876 2.154 0.0357 *---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3185 on 55 degrees of freedomMultiple R-squared: 0.2085, Adjusted R-squared: 0.1797 F-statistic: 7.242 on 2 and 55 DF, p-value: 0.001614$B_83Call:lm(formula = log10(x) ~ mergy$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.90287 -0.32839 0.03175 0.37027 0.81465 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) -0.73480 0.45228 -1.625 0.11mergy$Rearing_Temp 0.03350 0.02004 1.672 0.10Residual standard error: 0.4479 on 56 degrees of freedomMultiple R-squared: 0.04753, Adjusted R-squared: 0.03052 F-statistic: 2.794 on 1 and 56 DF, p-value: 0.1002$B_70Call:lm(formula = log10(x) ~ mergy$bio5 + mergy$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.80424 -0.20413 -0.03442 0.25526 0.81219 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -1.479502 0.641632 -2.306 0.0249 * mergy$bio5 -0.002799 0.001965 -1.424 0.1600 mergy$Rearing_Temp 0.097784 0.014196 6.888 5.75e-09 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3171 on 55 degrees of freedomMultiple R-squared: 0.4779, Adjusted R-squared: 0.4589 F-statistic: 25.17 on 2 and 55 DF, p-value: 1.734e-08$B_40Call:lm(formula = log10(x) ~ mergy$Rearing_Temp + mergy$Axis.2 + mergy$Axis.3)Residuals: Min 1Q Median 3Q Max -1.32627 -0.32412 0.04458 0.31258 0.90367 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -0.92089 0.50132 -1.837 0.071726 . mergy$Rearing_Temp 0.04213 0.02214 1.903 0.062428 . mergy$Axis.2 -2.51976 1.04941 -2.401 0.019819 * mergy$Axis.3 -6.14160 1.47835 -4.154 0.000117 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.4782 on 54 degrees of freedomMultiple R-squared: 0.3049, Adjusted R-squared: 0.2663 F-statistic: 7.897 on 3 and 54 DF, p-value: 0.00018522016-11-01 adding full models with automated stepAIC
apply(merg[,38:43],2,function(x){summary(stepAIC(lm(log10(x)~merg$bio5+merg$Rearing_Temp+merg$Axis.1+merg$Axis.2+merg$Axis.3)),direction="forward")})Start: AIC=-135.83log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.3 1 0.00006 4.2616 -137.82- merg$Axis.2 1 0.00563 4.2671 -137.75- merg$Axis.1 1 0.03032 4.2918 -137.42- merg$bio5 1 0.05267 4.3142 -137.12<none> 4.2615 -135.83- merg$Rearing_Temp 1 0.32622 4.5877 -133.62Step: AIC=-137.82log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 Df Sum of Sq RSS AIC- merg$Axis.2 1 0.00557 4.2671 -139.75- merg$Axis.1 1 0.03288 4.2944 -139.39- merg$bio5 1 0.05995 4.3215 -139.03<none> 4.2616 -137.82- merg$Rearing_Temp 1 0.32790 4.5895 -135.60Step: AIC=-139.75log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 Df Sum of Sq RSS AIC- merg$Axis.1 1 0.02927 4.2964 -141.36- merg$bio5 1 0.05486 4.3220 -141.02<none> 4.2671 -139.75- merg$Rearing_Temp 1 0.35722 4.6243 -137.17Step: AIC=-141.36log10(x) ~ merg$bio5 + merg$Rearing_Temp Df Sum of Sq RSS AIC- merg$bio5 1 0.02771 4.3241 -142.99<none> 4.2964 -141.36- merg$Rearing_Temp 1 0.33717 4.6336 -139.05Step: AIC=-142.99log10(x) ~ merg$Rearing_Temp Df Sum of Sq RSS AIC<none> 4.3241 -142.99- merg$Rearing_Temp 1 0.3481 4.6722 -140.58Start: AIC=-147.19log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.1 1 0.009107 3.5000 -149.05- merg$Axis.3 1 0.009894 3.5008 -149.03- merg$bio5 1 0.016701 3.5076 -148.92- merg$Axis.2 1 0.046939 3.5379 -148.43<none> 3.4909 -147.19- merg$Rearing_Temp 1 0.215627 3.7065 -145.78Step: AIC=-149.05log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.3 1 0.005260 3.5053 -150.96- merg$bio5 1 0.008554 3.5086 -150.91- merg$Axis.2 1 0.057491 3.5575 -150.12<none> 3.5000 -149.05- merg$Rearing_Temp 1 0.210727 3.7107 -147.71Step: AIC=-150.96log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.2 Df Sum of Sq RSS AIC- merg$bio5 1 0.006235 3.5115 -152.86- merg$Axis.2 1 0.059127 3.5644 -152.01<none> 3.5053 -150.96- merg$Rearing_Temp 1 0.218048 3.7233 -149.52Step: AIC=-152.86log10(x) ~ merg$Rearing_Temp + merg$Axis.2 Df Sum of Sq RSS AIC- merg$Axis.2 1 0.065809 3.5773 -153.80<none> 3.5115 -152.86- merg$Rearing_Temp 1 0.225290 3.7368 -151.31Step: AIC=-153.8log10(x) ~ merg$Rearing_Temp Df Sum of Sq RSS AIC<none> 3.5773 -153.8- merg$Rearing_Temp 1 0.18654 3.7639 -152.9Start: AIC=-122.77log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.3 1 0.01759 5.0640 -124.58- merg$Axis.1 1 0.03695 5.0833 -124.37- merg$bio5 1 0.09873 5.1451 -123.69- merg$Axis.2 1 0.14349 5.1899 -123.20<none> 5.0464 -122.77- merg$Rearing_Temp 1 0.61137 5.6577 -118.37Step: AIC=-124.58log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 Df Sum of Sq RSS AIC- merg$Axis.1 1 0.06171 5.1257 -125.90- merg$Axis.2 1 0.13474 5.1987 -125.11- merg$bio5 1 0.15531 5.2193 -124.89<none> 5.0640 -124.58- merg$Rearing_Temp 1 0.62522 5.6892 -120.06Step: AIC=-125.9log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.2 Df Sum of Sq RSS AIC- merg$bio5 1 0.11746 5.2431 -126.63- merg$Axis.2 1 0.17282 5.2985 -126.04<none> 5.1257 -125.90- merg$Rearing_Temp 1 0.66713 5.7928 -121.05Step: AIC=-126.63log10(x) ~ merg$Rearing_Temp + merg$Axis.2 Df Sum of Sq RSS AIC<none> 5.2431 -126.63- merg$Axis.2 1 0.21853 5.4617 -126.35- merg$Rearing_Temp 1 0.63456 5.8777 -122.23Start: AIC=-85.77log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.2 1 0.09471 10.350 -87.247- merg$bio5 1 0.14357 10.399 -86.979- merg$Axis.3 1 0.17560 10.431 -86.803- merg$Rearing_Temp 1 0.34221 10.597 -85.900<none> 10.255 -85.771- merg$Axis.1 1 0.51791 10.773 -84.963Step: AIC=-87.25log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$bio5 1 0.09885 10.449 -88.705- merg$Axis.3 1 0.20541 10.555 -88.127- merg$Rearing_Temp 1 0.28656 10.636 -87.690<none> 10.350 -87.247- merg$Axis.1 1 0.45249 10.802 -86.808Step: AIC=-88.71log10(x) ~ merg$Rearing_Temp + merg$Axis.1 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Rearing_Temp 1 0.30750 10.756 -89.052<none> 10.449 -88.705- merg$Axis.3 1 0.37408 10.823 -88.700- merg$Axis.1 1 0.60533 11.054 -87.495Step: AIC=-89.05log10(x) ~ merg$Axis.1 + merg$Axis.3 Df Sum of Sq RSS AIC<none> 10.756 -89.052- merg$Axis.3 1 0.42229 11.178 -88.857- merg$Axis.1 1 0.71553 11.472 -87.381Start: AIC=-122.03log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$bio5 1 0.0001 5.4282 -124.032- merg$Axis.2 1 0.0329 5.4610 -123.689- merg$Axis.1 1 0.0409 5.4690 -123.605- merg$Axis.3 1 0.0666 5.4947 -123.338<none> 5.4281 -122.033- merg$Rearing_Temp 1 4.5125 9.9406 -89.546Step: AIC=-124.03log10(x) ~ merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.2 1 0.0357 5.4639 -125.659- merg$Axis.3 1 0.0798 5.5080 -125.200- merg$Axis.1 1 0.1695 5.5977 -124.279<none> 5.4282 -124.032- merg$Rearing_Temp 1 4.5125 9.9407 -91.545Step: AIC=-125.66log10(x) ~ merg$Rearing_Temp + merg$Axis.1 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$Axis.3 1 0.0784 5.5423 -126.847- merg$Axis.1 1 0.1733 5.6372 -125.879<none> 5.4639 -125.659- merg$Rearing_Temp 1 4.5377 10.0016 -93.197Step: AIC=-126.85log10(x) ~ merg$Rearing_Temp + merg$Axis.1 Df Sum of Sq RSS AIC- merg$Axis.1 1 0.1750 5.7173 -127.075<none> 5.5423 -126.847- merg$Rearing_Temp 1 4.4787 10.0209 -95.087Step: AIC=-127.07log10(x) ~ merg$Rearing_Temp Df Sum of Sq RSS AIC<none> 5.7173 -127.075- merg$Rearing_Temp 1 4.7398 10.4571 -94.659Start: AIC=-78.04log10(x) ~ merg$bio5 + merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC- merg$bio5 1 0.1636 11.382 -79.225<none> 11.219 -78.036- merg$Axis.1 1 0.4666 11.685 -77.754- merg$Rearing_Temp 1 0.6847 11.903 -76.718- merg$Axis.2 1 0.9679 12.186 -75.402- merg$Axis.3 1 3.9432 15.162 -63.168Step: AIC=-79.23log10(x) ~ merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3 Df Sum of Sq RSS AIC<none> 11.382 -79.225- merg$Axis.1 1 0.4311 11.813 -79.144- merg$Rearing_Temp 1 0.6969 12.079 -77.897- merg$Axis.2 1 0.8346 12.217 -77.263- merg$Axis.3 1 3.9224 15.305 -64.643$FC_83Call:lm(formula = log10(x) ~ merg$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.61666 -0.14861 -0.03988 0.14529 0.74191 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.36231 0.28484 1.272 0.2087 merg$Rearing_Temp 0.02638 0.01254 2.104 0.0399 *---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.2804 on 55 degrees of freedomMultiple R-squared: 0.07451, Adjusted R-squared: 0.05768 F-statistic: 4.428 on 1 and 55 DF, p-value: 0.03995$FC_70Call:lm(formula = log10(x) ~ merg$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.6417 -0.1415 0.0238 0.1711 0.3910 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 2.06670 0.25908 7.977 9.51e-11 ***merg$Rearing_Temp -0.01931 0.01140 -1.694 0.096 . ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.255 on 55 degrees of freedomMultiple R-squared: 0.04956, Adjusted R-squared: 0.03228 F-statistic: 2.868 on 1 and 55 DF, p-value: 0.09601$FC_40Call:lm(formula = log10(x) ~ merg$Rearing_Temp + merg$Axis.2)Residuals: Min 1Q Median 3Q Max -0.80392 -0.10073 0.07339 0.22020 0.55569 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.09071 0.32917 0.276 0.7839 merg$Rearing_Temp 0.03680 0.01453 2.533 0.0143 *merg$Axis.2 1.24166 0.83541 1.486 0.1431 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3145 on 53 degrees of freedom (1 observation deleted due to missingness)Multiple R-squared: 0.166, Adjusted R-squared: 0.1345 F-statistic: 5.275 on 2 and 53 DF, p-value: 0.008145$B_83Call:lm(formula = log10(x) ~ merg$Axis.1 + merg$Axis.3)Residuals: Min 1Q Median 3Q Max -0.89374 -0.32249 0.03374 0.32440 0.77433 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.01073 0.05911 0.182 0.8566 merg$Axis.1 -1.09010 0.57516 -1.895 0.0634 .merg$Axis.3 2.00468 1.37680 1.456 0.1512 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.4463 on 54 degrees of freedomMultiple R-squared: 0.09566, Adjusted R-squared: 0.06217 F-statistic: 2.856 on 2 and 54 DF, p-value: 0.06621$B_70Call:lm(formula = log10(x) ~ merg$Rearing_Temp)Residuals: Min 1Q Median 3Q Max -0.7507 -0.1789 -0.0132 0.2067 0.7046 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -2.24217 0.32753 -6.846 6.75e-09 ***merg$Rearing_Temp 0.09734 0.01442 6.753 9.59e-09 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.3224 on 55 degrees of freedomMultiple R-squared: 0.4533, Adjusted R-squared: 0.4433 F-statistic: 45.6 on 1 and 55 DF, p-value: 9.589e-09$B_40Call:lm(formula = log10(x) ~ merg$Rearing_Temp + merg$Axis.1 + merg$Axis.2 + merg$Axis.3)Residuals: Min 1Q Median 3Q Max -1.38234 -0.22276 -0.00071 0.25240 0.84201 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -0.84687 0.49824 -1.700 0.09527 . merg$Rearing_Temp 0.03887 0.02200 1.767 0.08319 . merg$Axis.1 -0.85399 0.61446 -1.390 0.17062 merg$Axis.2 -2.42734 1.25523 -1.934 0.05870 . merg$Axis.3 -6.12416 1.46081 -4.192 0.00011 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.4724 on 51 degrees of freedom (1 observation deleted due to missingness)Multiple R-squared: 0.3278, Adjusted R-squared: 0.2751 F-statistic: 6.218 on 4 and 51 DF, p-value: 0.0003732Page 84: 2016-10-12]. Updating climate cascade to do list.
Project updates:
Hsp gene expression + Ctmax project:
- Go over updated figures
- Starting to write: working title-Shifts in the reaction norms of heat shock protein gene expression accompany evolutionary innovations in thermal tolerance of forest ants
Multiple stressors ms:
- Sent SHC another version
- Sent SHC another version
Range limits ms: SHC lab gave verbal edits:
- focus on 1 end of thermal niche breadth(although it is nice to mention it because CTmin decreases across lat)--CTmin.
- Dicussion needs to talk about cold adaptation; why trade-offs?
- Walk through results better
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- in reference to missing samples
- Fit in time to process Curtis' samples.
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: should be getting data soon
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Talk title: Northern range limits of a common forest ant is associated with trade-offs in cold physiology
- Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 85: 2016-10-14. Paper note: Puentes, A., G. Granath, and J. Ågren. 2016. Similarity in G matrix structure among natural populations of Arabidopsis lyrata. Evolution 70:2370–2386.
Similar paper here: Page 60: 2016-09-01. Paper notes: Paccard A, Van Buskirk J, Willi Y, Eckert CG, Bronstein JL. 2016. Quantitative Genetic Architecture at Latitudinal Range Boundaries: Reduced Variation but Higher Trait Independence. The American Naturalist.
Method differences:
But Puentes et al. focus on A. lyrata in thier native range(Norway-Sweden) and in the field, and Paccard et al. 2016 use populations from USA-Cananda in lab conditions. Peuntes uses 5 trais, Paccard used 10 traits. One of Cortlett Wood's papers suggests that the number of traits can alter how G changes among environments. Check what traits were used between studies.
Summary of findings: G is stable between Norway and Sweden populations
Page 86: 2016-10-14. Wiley House Style Guide
I'll need to follow these general writing rules for submitting a ms to Evolution.
Use of that and which
* That is used for defining or restrictive clauses: * The patient made a list of the symptoms that were most troublesome A defining clause is specific (limiting) to a particular person or thing; i.e., the patient had to list only those particular symptoms that were most troublesome. * Which is used in nondefining or nonrestrictive clauses: * The patient made a list of the symptoms, which were most troublesome A nondefining clause is general (nonlimiting); it provides additional information, and the use of commas is often important. In this example, all the symptoms were very troublesome.
Redundancy
Avoid using a modifying word when the intended meaning is inherent in a word already used. Redundancy is obvious in examples such as the results were plotted graphically, past history, bright blue in color, inactivates its activity, and completely filled. Does the term careful monitoring suggest that the alternative is careless monitoring?
Balancing a sentence:
It is important to ensure that a sentence balances on either side of certain words (correlatives) that emphasize similarity or contrast and that are used in parallel: both and and; either and or; neither and nor; not only and but; between and and;whether and or. For example,“I swam both in the morning and afternoon” should be “I swam both in the morning and in the afternoon”or “I swam in both the morning and the afternoon.”Note the position of the preposition in. (See also the section “Editing for Sense.”)
Key Points
• It is now acceptable to use the active or the passive voice. • Use the past tense for the author’s methods and results, and the present tense for interpretation and generally accepted “facts.” • The subject and verbmust agree in number. • “That” is defining;“which” is not. • Check that articles (“a,”“an,” and “the”) are used correctly. • Sentences must balance (e.g.,with “both … and …”). • In comparisons (e.g.,with lower/higher/less/more),make sure it is clear what is being compared with what. • Avoid sexist, dehumanizing, and stereotypical language.
Punctuation
Semi colons and Colons
SEMICOLONS • The semicolon is stronger than a comma but not as decisive as a full point. It can be used to separate sentences (whereas a comma cannot). • Use a semicolon before, and a comma after, the conjunctive adverbs however, that is, nevertheless, etc. COLONS Colons are used to introduce material that restates, explains, enlarges upon, or summarizes previous material. They also introduce items in a list set off from text (but a colon is not needed in run-on lists introduced by the words for example, namely, including, etc.; e.g., in the sentence “The dessert looks nice with fruit on it, for example: strawberries, raspberries, and blueberries” the colon should not be there). • In US spelling, if the material introduced by a colon consists of more than one sentence, or if it is a formal statement, quotation, or speech in dialogue it should take a capital after the colon. In UK spelling, a capital letter is not used after a colon (except in titles and subtitles). • Ratios containing words should have a thin space on each side of the colon (e.g., the light : dark cycle) but ratios containing numbers should be closed up (e.g., 16:8 h).
Key Points:
• Use commas to clarify sentences. • Do not use a comma to separate sentences; use a semicolon (this is a particularly common error before “however” and “nevertheless”). • Do not use apostrophes with plural abbreviations (e.g.,ANOVAs, not ANOVA’s). • For hyphenation, refer to your journal style sheet. • Do not hyphenate adverbs ending in -ly (e.g., dermatologically tested soap). • Use hyphens in compound terms to clarify meaning (e.g.,much-needed clothing). • Use en dashes, not hyphens, for associations (e.g., dose–response curve).
Page 87: 2016-10-14. NSF post doc app meeting: Keller Lab
SK background to grant
NSF use to have bioinformatics post doc competition and replaced with narrowly defined one in bio. It has to fit into 1 or a couple bins: 1 of them isplant genome research program (PGRP; funds poplar). SK attend PGRP meetings as part of training missions seriously. They build a program and come in as cohort(post doc fellows) and they ahve extra training sessions with them. Post doc presents work and are well supported for 3 years. SK fits squarely into: economically important plant, genome wide approaches to the problem of plant growth/yield and response to stress and other challenges.
Project Summary
Project description
Large communication issue
- What is new and novel? Kattia
- Figure 3: analysis is of a single trait Hammer it down, multiple times, outside of fig legend and make it more clear.
- HAMMER DOWN novelty is non-linear GxE interactions
- Cant predict performance readily from 1 environment to another environment (that span the current and future climate)
- Say you'll measure wood traits
- Bring more genomics more important: Bring in population genetics into the proposal.
- Add path analsysis
- Come up with precise alleles of what is adaptive.
- Fig 1 C. put an ellipse for central population:
- Set margins to 1 inch around.
- heavy lifting (SK): bring emphasis on gene expression way up (genetic variation among genotpes in their transcriptional response to that variation); ID genes or networks of genes that show differences in expression or organization. What parts of the transcription? GO, pathways? Genes in trade-offs in few networks or overdispered across a network, relative to the total transcriptome. Stress response genes (Hsps)? Phenology associated genes (circadian clock). How can that be pulled out using the kingsolver method. (Not just as a tool that is cool to use, but as a question with an appropriately matched tool).
- Look at the SNPs. include in
- There is gxp from fairbanks and indian head. "Timing for success title"
- Karl: pair down first paragraph; reduce in length
- SK, focus on the major ideas
- Be more explicit about what the trait is used for Gmatrix.
- genetically based differences to identify GxE
Dissertation Abstract
Data management
Page 88: 2016-10-18. Climate cascade meeting
Project updates:
Hsp gene expression + Ctmax project:
- Go over updated figures
- figure 3, SHC says to switch back branches
- figure 4, color code by habitat type, NJG:don't use dot dash, use dash
- Starting to write: working title-Shifts in the reaction norms of heat shock protein gene expression accompany evolutionary innovations in thermal tolerance of forest ants
- need to start writing methods and results
- Go over updated figures
Multiple stressors ms:
- Sent SHC another version ; should submit soon
- Sent SHC another version ; should submit soon
Range limits ms: SHC lab gave verbal edits:
- focus on 1 end of thermal niche breadth(although it is nice to mention it because CTmin decreases across lat)--CTmin.
- Dicussion needs to talk about cold adaptation; why trade-offs?
- Walk through results better
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project: should be getting data soon
Attending SICB - Jan 4-8 New Orleans, Give a talk about range limits paper.
Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
Talk title: Northern range limits of a common forest ant is associated with trade-offs in cold physiology
Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
- Application submitted today 2016-10-18
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 89: 2016-10-25. Climate cascade updated list
Project updates:
Hsp gene expression + Ctmax project:
- figure 3, SHC says to switch back branches
- Starting to write: working title-Shifts in the reaction norms of heat shock protein gene expression accompany evolutionary innovations in thermal tolerance of forest ants
- need to start writing methods and results; submit to MBE
Multiple stressors ms:
- submitted 2016-10-24
- submitted 2016-10-24
Range limits ms: SHC lab gave verbal edits:
- focus on 1 end of thermal niche breadth(although it is nice to mention it because CTmin decreases across lat)--CTmin.
- Dicussion needs to talk about cold adaptation; why trade-offs?
- Walk through results better
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Modulation of Hsp ms:
- make fig 2 without spline curves with just points
- grab elevation data for each sampling point in R
- make fig 2 without spline curves with just points
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
Talk title: Northern range limits of a common forest ant is associated with trade-offs in cold physiology
Apply for funding. Suitor Travel Grant Deadline is october 31
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
- Application submitted today 2016-10-18
- Wrote up suiter award app. I need to find out pricing (~ $1000) and then get everything signed. Waiting to find better flight prices.
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Dissertation Abstract is in multiple paragraphs, but for dissertation itself, make 1 paragraph
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 90: 2016-10-25. Meeting with M Pespeni
Meeting time, Wednesday 2-4; 2016-10-26
Things to discuss
Potential post doc oppportunity at MBL(Marine Biological Laboratory)
- Previous email pitch with prospective post doc mentor
- Previous email pitch with prospective post doc mentor
A question that excites me is how organisms persist and respond to environmental change in natural populations? ( This falls into 2 strategic themes of MBL: comparative evolution and genomics, and organismal adaptation and resiliency to climate change) Well, response to selection depends on their quantitative genetic architecture (variance-covariance G matrix) and selection gradient. Monogonont rotifers seem like a really great system to explore this question with a combination of field surveys, and lab studies. For example, since their lifespan is relatively short, there should be a lot of evolutionary responses within a season. So this would involve sampling rotifers throughout the season (4-6 times), then genotyping (GBS or rad-seq, maybe whole genome sequencing if its not too large) and establishing clones each time. Genotyping would detect shifts in allele frequencies with respect to the environment that changes, signature of evolution. Establishing clones would allow one to assess the evolutionary potential at each point in the season by estimating the variance-covariance G matrix. Selection should erode genetic variation, so G should be altered throughout the season that may hinder or facilitate future responses. And evolutionary potential is really unique in rotifers because they can be clonal or mate. So I'd be interested in comparing G between these life strategies. The problem with a G matrix, is that we have no clue what the key molecular players are: so to tackle this problem, one could leverage the collected data into a qtl analysis too.
I think it is fun to think about the evolutionary potential to environmental change for organisms that can switch from asexual to sexual reproduction. If you compare the G matrix between them, sexually produced offspring populations should have more genetic variance than clonal offspring populations. These animals are resilient to environmental change because of this! So it'd be cool to compare G between asexual vs sexual and whether trade-offs can shift among traits.
Melissa advice; write down questions, hypotheses and aims that will help facilitate the discussion
- sequencing for ecological genomics? : multiepex individuals , you'll need 1-2x converage: or pool individuals and estimate allele frequencies (sequence RNA or DNA); if RNA, then you'll have potential for allele-biased expression influencing allele freq estimates. If DNA gnoemics from a pooled sample, then playing field is level, but genomes are big. 2 ways to do it: HARP(genotype parentals(known)-then subsequent genotype larvae; needs and requires low coverage---then reconstruct allele frequencies).
- How many individuals per pop (10-100?) depends on how large your pop size (only need a few individuals)? If small pop--need more and there will be more random chance. Look up Christian Slaughter (experimental evolution). Look up papers ; power analyses.
- GTA for ecological genomics
2016-10-27 Brent's thoughts
Ask about Isofemale lines
- look up genome size (it's .35 pg)
- What is changing G? What is the predictive power? model it.
- Try to talk to Mike Angiletta, Rus Lande. (Genetic accomodation and assimilation)
Page 91: 2016-10-26 SICB meeting talk
125-7 Sunday, Jan. 8 11:45
Page 92: 2016-10-27. Proteome stability project update
reminder: generated unfolding reaction norms for 6 ant colonies (3 colonies per species).
received data from Bethany 2016-10-26
excel sheets wit relative abundance to first sample is in: 2016Protein_stability_evolution/Data/2016/10Oct
- in this path, you'll see 3 folders, 1 set of samples queried against 18 species (it actually has a combo of ants/microbes because Bethany just took the top 18 searches) from uniprot. The other folder queries the NCBI database. And the last folder contains raw mass spec files.
- in this path, you'll see 3 folders, 1 set of samples queried against 18 species (it actually has a combo of ants/microbes because Bethany just took the top 18 searches) from uniprot. The other folder queries the NCBI database. And the last folder contains raw mass spec files.
Bethany is going to run more of the sample to see if we can ID more proteins.
Page 93: 2016-10-31. CTmax and Hsp reaction norm stats
Stats overview:
- Effect of local environment (Tmax and habitat type) on basal xp and other parameters.
Basal xp
summary(lm(b70~bio5+habitat_v2,data=b70))Call:lm(formula = b70 ~ bio5 + habitat_v2, data = b70)Residuals: Min 1Q Median 3Q Max -2.10674 -0.34255 0.07049 0.44475 1.56186 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 10.915110 1.957855 5.575 1.74e-06 ***bio5 0.005714 0.006543 0.873 0.388 habitat_v2flat woods -0.124177 0.365522 -0.340 0.736 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.87 on 41 degrees of freedomMultiple R-squared: 0.01836, Adjusted R-squared: -0.02952 F-statistic: 0.3835 on 2 and 41 DF, p-value: 0.6839summary(lm(b83~bio5+habitat_v2,data=b83))Call:lm(formula = b83 ~ bio5 + habitat_v2, data = b83)Residuals: Min 1Q Median 3Q Max -2.16408 -0.49336 0.03001 0.64313 1.96466 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 12.751247 2.132030 5.981 2.34e-07 ***bio5 -0.002689 0.007140 -0.377 0.708 habitat_v2flat woods -0.480410 0.362781 -1.324 0.191 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.9811 on 50 degrees of freedomMultiple R-squared: 0.06047, Adjusted R-squared: 0.02289 F-statistic: 1.609 on 2 and 50 DF, p-value: 0.2103summary(lm(b40~bio5+habitat_v2,data=b40))Call:lm(formula = b40 ~ bio5 + habitat_v2, data = b40)Residuals: Min 1Q Median 3Q Max -1.7137 -0.6858 -0.1241 0.3196 3.0774 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 13.721471 2.714855 5.054 1.36e-05 ***bio5 0.004703 0.009120 0.516 0.609 habitat_v2flat woods -0.381890 0.509208 -0.750 0.458 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.123 on 35 degrees of freedomMultiple R-squared: 0.01669, Adjusted R-squared: -0.0395 F-statistic: 0.297 on 2 and 35 DF, p-value: 0.7449Hsp70 (hsc70-4 h2) params (slope,Tm,max)
apply(b[,3:5],2,function(x){summary(lm(x~b$bio5+b$habitat_v2))})$FC_hsc70_1468_maxCall:lm(formula = x ~ b$bio5 + b$habitat_v2)Residuals: Min 1Q Median 3Q Max -20.536 -8.414 -1.652 4.839 30.045 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -0.90802 28.24097 -0.032 0.974489 b$bio5 0.13378 0.09449 1.416 0.163567 b$habitat_v2flat woods 20.35661 4.86449 4.185 0.000127 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 12.34 on 46 degrees of freedomMultiple R-squared: 0.4224, Adjusted R-squared: 0.3973 F-statistic: 16.82 on 2 and 46 DF, p-value: 3.288e-06$FC_hsc70_1468_slopeCall:lm(formula = x ~ b$bio5 + b$habitat_v2)Residuals: Min 1Q Median 3Q Max -0.91667 -0.22656 0.08771 0.27554 0.87662 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.213328 1.023087 0.209 0.83575 b$bio5 0.002091 0.003423 0.611 0.54431 b$habitat_v2flat woods 0.494706 0.176226 2.807 0.00731 **---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.4471 on 46 degrees of freedomMultiple R-squared: 0.2228, Adjusted R-squared: 0.189 F-statistic: 6.595 on 2 and 46 DF, p-value: 0.003032$FC_hsc70_1468_TmCall:lm(formula = x ~ b$bio5 + b$habitat_v2)Residuals: Min 1Q Median 3Q Max -2.23057 -0.46633 -0.00151 0.62405 1.24574 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 35.043684 1.956972 17.907 < 2e-16 ***b$bio5 0.003766 0.006548 0.575 0.568014 b$habitat_v2flat woods 1.372953 0.337088 4.073 0.000181 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8552 on 46 degrees of freedomMultiple R-squared: 0.3566, Adjusted R-squared: 0.3287 F-statistic: 12.75 on 2 and 46 DF, p-value: 3.931e-05Hsp83 params (slope,Tm,max)
apply(u[,9:11],2,function(x){summary(lm(x~u$bio5+u$habitat_v2))})$FC_Hsp83_279_maxCall:lm(formula = x ~ u$bio5 + u$habitat_v2)Residuals: Min 1Q Median 3Q Max -7.8432 -2.7507 -0.7032 2.3143 11.2074 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 8.059606 8.941439 0.901 0.37208 u$bio5 -0.002729 0.029897 -0.091 0.92766 u$habitat_v2flat woods 4.720030 1.550712 3.044 0.00386 **---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 4.06 on 46 degrees of freedomMultiple R-squared: 0.2054, Adjusted R-squared: 0.1709 F-statistic: 5.947 on 2 and 46 DF, p-value: 0.005045$FC_Hsp83_279_slopeCall:lm(formula = x ~ u$bio5 + u$habitat_v2)Residuals: Min 1Q Median 3Q Max -1.8619 -0.5948 0.1370 0.6879 1.3637 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) -1.056652 1.865514 -0.566 0.574u$bio5 0.008211 0.006238 1.316 0.195u$habitat_v2flat woods 0.301698 0.323536 0.933 0.356Residual standard error: 0.8471 on 46 degrees of freedomMultiple R-squared: 0.09876, Adjusted R-squared: 0.05957 F-statistic: 2.52 on 2 and 46 DF, p-value: 0.09148$FC_Hsp83_279_TmCall:lm(formula = x ~ u$bio5 + u$habitat_v2)Residuals: Min 1Q Median 3Q Max -4.4767 -0.7621 0.1731 0.9167 2.6581 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 31.80124 3.37214 9.431 2.54e-12 ***u$bio5 0.01076 0.01128 0.955 0.344699 u$habitat_v2flat woods 2.16554 0.58483 3.703 0.000569 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.531 on 46 degrees of freedomMultiple R-squared: 0.3423, Adjusted R-squared: 0.3137 F-statistic: 11.97 on 2 and 46 DF, p-value: 6.533e-05Hsp40 (hsc70-4 h2) params (slope,Tm,max)
apply(n[,6:8],2,function(x){summary(lm(x~n$bio5+n$habitat_v2))})$FC_hsp40_541_maxCall:lm(formula = x ~ n$bio5 + n$habitat_v2)Residuals: Min 1Q Median 3Q Max -7.8615 -3.3291 -0.6736 1.7653 10.5454 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 9.4754213 10.9534314 0.865 0.3917 n$bio5 -0.0009401 0.0367220 -0.026 0.9797 n$habitat_v2flat woods 3.6490726 1.8969491 1.924 0.0609 .---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 4.749 on 44 degrees of freedomMultiple R-squared: 0.1003, Adjusted R-squared: 0.05945 F-statistic: 2.454 on 2 and 44 DF, p-value: 0.09765$FC_hsp40_541_slopeCall:lm(formula = x ~ n$bio5 + n$habitat_v2)Residuals: Min 1Q Median 3Q Max -1.4300 -0.5157 0.2182 0.6412 1.3309 Coefficients: Estimate Std. Error t value Pr(>|t|)(Intercept) -0.295834 1.816631 -0.163 0.871n$bio5 0.005677 0.006090 0.932 0.356n$habitat_v2flat woods 0.413173 0.314610 1.313 0.196Residual standard error: 0.7877 on 44 degrees of freedomMultiple R-squared: 0.1048, Adjusted R-squared: 0.06411 F-statistic: 2.576 on 2 and 44 DF, p-value: 0.08755$FC_hsp40_541_TmCall:lm(formula = x ~ n$bio5 + n$habitat_v2)Residuals: Min 1Q Median 3Q Max -3.7066 -1.0076 0.2038 0.9873 3.5691 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 39.14520 3.84815 10.172 3.93e-13 ***n$bio5 -0.01175 0.01290 -0.911 0.367444 n$habitat_v2flat woods 2.46904 0.66643 3.705 0.000588 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.669 on 44 degrees of freedomMultiple R-squared: 0.2539, Adjusted R-squared: 0.22 F-statistic: 7.487 on 2 and 44 DF, p-value: 0.00159Summary: no sig effect of Tmax (bio5) on parameters, but habitat type does in some cases:
Table summary:
| Parameter | hsp83 | hsc70.4.h2 | hsp40 |
|---|---|---|---|
| basal | no | no | no |
| slope | no | yes | no |
| Tm | yes | yes | yes |
| max | yes | yes | no |
Page 94: 2016-10-31; 2016-11-01. Climate cascade meeting setup and notes
Project updates:
Hsp gene expression + Ctmax project:
- figure 3, SHC says to switch back branches
- Wrote up methods and results
- Submit to? MBE, evolution, Goerge Somero and Brent think PNAS is a good fit. SHC and NJG thoughts?
- reference for rad-seq:HF3-picea,fbragg2-floridana,KH4-ashmeadi,Duke6-mariae,ala2-miamiana, Lex13-rudis
Multiple stressors ms:
- submitted 2016-10-24
- in review 2016-11-01
- submitted 2016-10-24
Range limits ms: SHC lab gave verbal edit, still need to incorporate
Thermal niche ms: Lacey and I working on discussion
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Modulation of Hsp ms:
- make fig 2 without spline curves with just points (done)
- grab elevation data for each sampling point in R (done)
- make fig 2 without spline curves with just points (done)
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
Apply for funding. Suitor Travel Grant Deadline is october 31
Wrote up suiter award app Application submitted today 2016-10-18
- Bought hotel, rooming with Emily M., need to buy airplane tickets
Thesis related FORMS FOUND HERE
Formatting:
- Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 95: 2016-11-02. Ancestral trait reconstruction and CTmax PGLS ANBE common garden
Ancestral trait reconstruction
cols<-ifelse(esthab[,1]> esthab[,2],"blue","red")par(mar=c(1,1,1,1))plot(ult.tree1,cex=.5)nodelabels(pch=19,cex=.75,col=cols)#obj<-contMap(ult.tree1,trait,plot=FALSE,fsize=.1,method="fastAnc")#obj$cols[]<-obj<-setMap(obj,colors=colorRampPalette(c("black","gray","red"))(length(obj$cols)))plot(obj,legend=FALSE)nodelabels(pch=19,cex=3,col=cols)
Using ancTHRESH: Paper here;troubleshooting error
Species level ancestral state reconstruction
####using ancThresh (revell 2014 evolution)habitat<-as.character(sm.dat2$Habitat)names(habitat)<-spec.tree$tip.labeler<-ancThresh(spec.tree,habitat,model="BM",ngen=20000)$ace DF FW9 0.9411765 0.0588235310 1.0000000 0.0000000011 0.9411765 0.0588235312 1.0000000 0.0000000013 1.0000000 0.0000000014 1.0000000 0.0000000015 0.1176471 0.88235294$mcmc 9 10 11 12 13 14 151 FW FW FW FW FW FW FW2 DF DF DF DF DF DF FW3 DF DF DF DF DF DF FW4 DF DF DF DF DF DF FW5 FW DF FW DF DF DF FW6 DF DF DF DF DF DF FW7 DF DF DF DF DF DF FW8 DF DF DF DF DF DF FW9 DF DF DF DF DF DF FW10 DF DF DF DF DF DF FW11 DF DF DF DF DF DF FW12 DF DF DF DF DF DF DF13 DF DF DF DF DF DF FW14 DF DF DF DF DF DF DF15 DF DF DF DF DF DF FW16 DF DF DF DF DF DF FW17 DF DF DF DF DF DF FW18 DF DF DF DF DF DF FW19 DF DF DF DF DF DF FW20 DF DF DF DF DF DF FW21 DF DF DF DF DF DF FW$par gen DF FW logLik1 0 0 Inf -20.4474772 1000 0 Inf -15.4975093 2000 0 Inf -7.4549564 3000 0 Inf -7.5834055 4000 0 Inf -12.4439486 5000 0 Inf -7.8246427 6000 0 Inf -18.4112818 7000 0 Inf -8.9576099 8000 0 Inf -10.36672010 9000 0 Inf -16.03234611 10000 0 Inf -10.06403412 11000 0 Inf -12.23285213 12000 0 Inf -11.15029214 13000 0 Inf -10.57509215 14000 0 Inf -11.40625316 15000 0 Inf -18.84279517 16000 0 Inf -13.00144118 17000 0 Inf -10.96166219 18000 0 Inf -10.05459620 19000 0 Inf -13.25141721 20000 0 Inf -13.367340$liab ashmeadi floridana picea rudis miamiana lamellidens1 0.4256323 0.41631348 -0.9982783 -0.7611795 -0.2870708 -0.72283742 1.7161387 2.01928329 -0.5807896 -2.7040955 -2.1537732 -1.38790363 0.2283514 0.33912611 -1.4167395 -0.8156619 -2.1513080 -1.56261124 0.2245830 0.04840176 -0.1672812 -0.5182768 -1.4955228 -1.28459575 2.8412873 2.52791039 -0.9945787 -0.1217164 -1.0169444 -0.81994816 0.1611044 0.07884604 -1.6873462 -1.9551489 -2.5062990 -1.87355457 0.6062956 0.67119993 -1.7010454 -3.1098352 -3.5942080 -3.65994008 0.2781314 0.90142051 -0.9775805 -1.4564663 -2.0262664 -1.99556509 0.4831741 0.32616809 -1.2045168 -1.4714718 -1.8546322 -1.944372010 0.9545092 0.91442789 -1.9678349 -2.8803130 -2.0902628 -2.142006611 0.5334539 0.45214518 -0.6975138 -1.7053550 -1.0307576 -1.367155512 0.3455715 0.36069694 -0.8319524 -1.3187262 -0.3870102 -0.677815513 1.0041606 0.14710811 -1.5117681 -1.1249621 -2.0614504 -1.651554314 0.3898680 0.07794064 -2.5767746 -2.2195374 -2.0482449 -2.531143315 0.1438718 0.01582491 -0.6168032 -1.8867342 -2.1514162 -2.211689316 1.5174549 1.25039739 -0.3146283 -0.6646803 -2.9459244 -2.406532717 0.2949082 0.79497182 -2.3475672 -1.3544484 -1.7933900 -1.063316818 0.1754723 0.07905511 -1.7017423 -2.8025226 -2.3548623 -2.776637619 0.6090604 0.62077613 -2.8309318 -2.2609481 -2.4802131 -2.922959920 0.3999724 0.84360557 -3.1013779 -2.7228427 -3.7007126 -3.268761421 0.7346061 0.86140836 -2.1879653 -2.5777420 -3.3467673 -3.9166340 fulva tennesseensis 9 10 11 121 -0.1323211 -0.4694033 0.1077790 0.05401888 0.1439697 0.55491072 -1.3488986 -0.9985338 -1.0447443 -0.89772529 -1.3726393 -1.62553533 -1.0540075 -0.8284915 -0.2338892 -1.07612349 -0.5740115 -1.02164614 -0.8706067 -1.3873548 -0.4220714 -1.45708985 -0.1885166 -0.48265535 -0.4714702 -0.3575748 0.3448237 -0.79814696 0.4179758 -0.11062866 -2.0032554 -1.5914376 -1.1376738 -1.68649095 -0.9229725 -1.57367727 -2.8018002 -3.0551338 -1.8844310 -2.27322487 -1.7170136 -2.40810148 -2.3101088 -2.1159034 -1.4001121 -2.09323137 -1.4599777 -1.83636679 -2.9641505 -2.4055435 -1.0906425 -2.44676880 -0.8556195 -0.370800310 -0.8302079 -1.8660746 -1.5512891 -1.57882060 -0.9111552 -0.784013911 -1.1688755 -0.5233950 -1.0694070 -1.12396533 -1.3927681 -0.875806512 -0.8464924 -1.0075126 -1.4279378 -1.32894559 -0.8023390 -1.166729213 -0.5218728 -0.5705261 -0.0381062 -0.92343185 -0.4744984 -1.305807914 -0.6052192 -0.3901746 -1.3237021 -0.60220033 -0.6998386 -1.692903715 -1.2232572 -1.3633033 -0.3002129 -1.09482578 -1.0404010 -0.815608816 -1.6465250 -2.5813912 -1.9776983 -2.28317185 -2.0259641 -1.165865717 -3.3390536 -3.0821085 -1.7921216 -3.21916257 -2.0360532 -2.629006318 -2.8353118 -2.5476584 -2.4669142 -2.85649615 -2.2842481 -2.431180619 -2.2636787 -2.3547350 -1.7062219 -1.86053981 -1.6758183 -1.944894820 -2.2528318 -2.1913204 -1.6877972 -1.97671417 -2.1753948 -2.638980121 -0.9965233 -0.5414665 -0.4247154 -0.76066224 -0.6674432 -1.9868298 13 14 151 0.9519656 0.7427504 0.240052462 -1.3613162 -1.6237741 1.618247213 -1.0603488 -1.7249034 0.510796674 -0.5200048 -1.3974409 0.287239045 -0.4768141 -1.2742356 2.251258966 -2.3110323 -2.0833774 0.160072297 -3.6709435 -3.6396813 0.983934238 -1.9847607 -2.2242295 0.690427149 -1.3939044 -1.9765742 0.3111988010 -1.5328568 -1.8498664 1.1619979211 -1.5364744 -1.0508674 0.5006389512 -0.3373547 -0.1329506 -0.1625127013 -1.2442089 -1.5792052 0.1002019914 -1.6824202 -2.2044414 -0.0460002815 -1.7298317 -2.0001838 0.5517434916 -1.4318329 -2.2727758 1.0112304117 -2.2424728 -1.5377536 0.3171352218 -2.8320154 -2.1532767 0.2184941319 -2.2843315 -2.4451599 0.4856873120 -3.4562176 -3.2262318 0.2699920721 -2.8093678 -3.7144363 0.53795324It automatically plots the results:

Reference for tree with node labels

Doing pgls in 3 ways:
- Using colonies as tips (breaks assumptions because of reticulate evolution)
- Forcing polytomies with species as replicates
- Just doing species themselves (8)
1. Using colonies as tips (breaks assumptions because of reticulate evolution)
library(caper)aph_phylo1$colony.id2<-as.character(aph_phylo1$colony.id2)ult.tree1<-makeLabel(ult.tree1)aph_phylo1$habitat_v2<-droplevels(aph_phylo1$habitat_v2)pp<-comparative.data(phy=ult.tree1,data=aph_phylo1,names.col=colony.id2, vcv = TRUE, na.omit = FALSE, warn.dropped = TRUE)momo<-pgls(KO_temp_worker~bio5+habitat_v2,data=pp,lambda="ML",bounds=list(lambda=c(0.001,1)))summary(momo)Call:pgls(formula = KO_temp_worker ~ bio5 + habitat_v2, data = pp, lambda = "ML", bounds = list(lambda = c(0.001, 1)))Residuals: Min 1Q Median 3Q Max -2.3636 -0.6161 -0.1511 0.3177 2.8311 Branch length transformations:kappa [Fix] : 1.000lambda [ ML] : 0.001 lower bound : 0.001, p = 1 upper bound : 1.000, p = < 2.22e-16 95.0% CI : (NA, 0.517)delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 37.2440770 1.0902152 34.1621 < 2.2e-16 ***bio5 0.0128318 0.0036686 3.4978 0.0007098 ***habitat_v2flat woods 1.3750216 0.2575557 5.3387 6.157e-07 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8605 on 97 degrees of freedomMultiple R-squared: 0.4051, Adjusted R-squared: 0.3929 F-statistic: 33.03 on 2 and 97 DF, p-value: 1.147e-11It looks like the PGLS is using lambda of 0. So I tried estimating lambda and then plugging it in the PGLS model
#phylogenetic signalx<-aph_phylo1$KO_temp_workernames(x)<-aph_phylo1$colony.id2phylosig(ult.tree1,x,test=TRUE,method="lambda")$lambda[1] 0.4833368$logL[1] -128.4395$logL0[1] -151.6493$P[1] 9.5454e-12#phylosig(ult.tree1,x,test=TRUE,method="K",nsim=1000)#redoing pgls with lambda from phylosigmomo3<-pgls(KO_temp_worker~habitat_v2+bio5,data=pp,lambda=0.4833368)summary(momo3)Call:pgls(formula = KO_temp_worker ~ habitat_v2 + bio5, data = pp, lambda = 0.4833368)Residuals: Min 1Q Median 3Q Max -2.3928 -0.3833 0.1074 0.8404 3.3408 Branch length transformations:kappa [Fix] : 1.000lambda [Fix] : 0.483delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 38.4681831 2.4673203 15.5911 <2e-16 ***habitat_v2flat woods 0.5009582 0.5160753 0.9707 0.3341 bio5 0.0093601 0.0080294 1.1657 0.2466 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.082 on 97 degrees of freedomMultiple R-squared: 0.02726, Adjusted R-squared: 0.007207 F-statistic: 1.359 on 2 and 97 DF, p-value: 0.2617 2. Forcing polytomies with species as replicates
aph_phylo2$colony.id2<-as.character(aph_phylo2$colony.id2)ult2.tree<-makeLabel(ult2.tree)aph_phylo2$habitat_v2<-droplevels(aph_phylo2$habitat_v2)pp<-comparative.data(phy=ult2.tree,data=aph_phylo2,names.col=colony.id2, vcv = TRUE, na.omit = FALSE, warn.dropped = TRUE)momo<-pgls(KO_temp_worker~bio5+habitat_v2,data=pp,lambda="ML",bounds=list(lambda=c(0.001,1)))summary(momo)Call:pgls(formula = KO_temp_worker ~ bio5 + habitat_v2, data = pp, lambda = "ML", bounds = list(lambda = c(0.001, 1)))Residuals: Min 1Q Median 3Q Max -5.2426 -1.0208 -0.0880 0.9807 5.7995 Branch length transformations:kappa [Fix] : 1.000lambda [ ML] : 0.991 lower bound : 0.001, p = 0.1627 upper bound : 1.000, p = 0.69339 95.0% CI : (NA, NA)delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 38.9471677 2.2565730 17.2594 <2e-16 ***bio5 0.0080535 0.0067643 1.1906 0.2367 habitat_v2flat woods -0.0036539 0.4282500 -0.0085 0.9932 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.839 on 97 degrees of freedomMultiple R-squared: 0.01442, Adjusted R-squared: -0.005903 F-statistic: 0.7095 on 2 and 97 DF, p-value: 0.4944 Again, try to estimate lambda and then plug and chug
x<-aph_phylo2$KO_temp_workernames(x)<-aph_phylo2$colony.id2phylosig(ult2.tree,x,test=TRUE,method="lambda")$lambda[1] 0.9759065$logL[1] -124.9107$logL0[1] -151.6493$P[1] 2.616073e-13momo3<-pgls(KO_temp_worker~habitat_v2+bio5,data=pp,lambda=0.9759065)summary(momo3)Call:pgls(formula = KO_temp_worker ~ habitat_v2 + bio5, data = pp, lambda = 0.9759065)Residuals: Min 1Q Median 3Q Max -3.9077 -1.1334 0.0055 1.0044 5.3637 Branch length transformations:kappa [Fix] : 1.000lambda [Fix] : 0.976delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 39.2879592 2.4115458 16.2916 <2e-16 ***habitat_v2flat woods 0.0118194 0.4401407 0.0269 0.9786 bio5 0.0069203 0.0073845 0.9371 0.3510 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 1.766 on 97 degrees of freedomMultiple R-squared: 0.009022, Adjusted R-squared: -0.01141 F-statistic: 0.4416 on 2 and 97 DF, p-value: 0.6443 3. Just doing species themselves (8)
#PGLS with caperspec.tree<-makeLabel(spec.tree)smp<-comparative.data(phy=spec.tree,data=sm.dat2,names.col=Species, vcv = TRUE, na.omit = FALSE, warn.dropped = TRUE)spmod<-pgls(CTmax~Habitat+Tmax,data=smp,lambda="ML")#spmod<-pgls(CTmax~Habitat,data=smp,lambda=0.885536,bounds=list(lambda=c(0.001,1)))summary(spmod)Call:pgls(formula = CTmax ~ Habitat + Tmax, data = smp, lambda = "ML")Residuals: Min 1Q Median 3Q Max -0.7707 -0.1147 0.0567 0.3244 0.5081 Branch length transformations:kappa [Fix] : 1.000lambda [ ML] : 0.000 lower bound : 0.000, p = 1 upper bound : 1.000, p = 0.0072118 95.0% CI : (NA, 0.738)delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 37.500342 2.914324 12.8676 5.048e-05 ***HabitatFW 1.462473 0.435376 3.3591 0.02013 * Tmax 0.011812 0.009520 1.2407 0.26975 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.4878 on 5 degrees of freedomMultiple R-squared: 0.7947, Adjusted R-squared: 0.7125 F-statistic: 9.674 on 2 and 5 DF, p-value: 0.01911 profile_lambda=pgls.profile(spmod, which="lambda") plot(profile_lambda)n<-sm.dat2$CTmaxnames(n)<-sm.dat2$Speciesphylosig(spec.tree,n,method="lambda",test=TRUE)$lambda[1] 0.885536$logL[1] -8.958222$logL0[1] -10.06035$P[1] 0.1376303spmod<-pgls(CTmax~Habitat+Tmax,data=smp,lambda=0.885536)summary(spmod)Call:pgls(formula = CTmax ~ Habitat + Tmax, data = smp, lambda = 0.885536)Residuals: Min 1Q Median 3Q Max -0.93013 -0.02903 0.07964 0.38357 1.47947 Branch length transformations:kappa [Fix] : 1.000lambda [Fix] : 0.886delta [Fix] : 1.000Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 39.3419794 4.4119919 8.9171 0.0002954 ***HabitatFW 1.6291565 0.9278322 1.7559 0.1394628 Tmax 0.0055482 0.0145875 0.3803 0.7193135 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8271 on 5 degrees of freedomMultiple R-squared: 0.467, Adjusted R-squared: 0.2538 F-statistic: 2.19 on 2 and 5 DF, p-value: 0.2074 Page 96: 2016-11-03. notes from skype meeting with KG, potential post doc opp
Marine Biological Labs, Hibbitt Early Career Fellows Program
about fellowship: its brand new for MBL; its trying to bring in new investigators early in their careers
MBL; resident and visiting scientists; there are a lot of courses in the summer (10 days to 6 weeks); teachers come from all of over the world; Whitman scholars are fellowships that PIs can establish labs; groups of researchers meet here;
Other foundations:
- Charles King foundation/trust
- Life sciences research foundation
- Ford Foundation
- Hell and Hay whitney foundation? check on deadlines
There is a genome for 15 different species. huge range in genome sizes, why?
Bioinformatics; david, mark welsh (bay paul center); Lots of peopel do pool-seq; own Illumina hi-seq ; miseq; sanger sequencing. play up bioinformatics resource;
MBL are conveners; convening power
Page 97: 2016-11-04. ms in prep
first authored
- multiple stressors (submitted)
- Curtis, stress in nature; submit to functional ecology
- rxn norm of Hsps and CTmax; submit to PNAS
- range limits paper with Jordan and Megan ; submit to American Naturalist
- Modulation of Hsp ms (in review)
- Proteome stability project (a stretch...)
with collaborators
- Comparative ramp papers (CP lead?); submit to current biology?
- (co-lead author) thermal niche paper with LChick; submit molecular ecology?
- CNP work with katie miller (submit where? )
Page 98: 2016-11-08. climate cascade meeting
Project updates:
Hsp gene expression + Ctmax project:
- figure 3, SHC says to switch back branches
- Wrote up methods and results-- go over with Nick then send to SHC
- Submit to PNAS
Multiple stressors ms:
- submitted 2016-10-24
- in review 2016-11-01
- submitted 2016-10-24
Range limits ms: SHC lab gave verbal edit, still need to incorporate
Thermal niche ms: Lacey and I working on discussion...eta?
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
Apply for funding. Suitor Travel Grant Deadline is october 31
Wrote up suiter award app Application submitted today 2016-10-18
- Bought hotel, rooming with Emily M., airplane tickets
Thesis related FORMS FOUND HERE
Formatting:
Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- started outline
- started outline
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 99: 2016-11-08. writing session with NJG
Writing Hsp reaction norm + CTmax ms in PNAS format
Someting to explore: variance among colony level means of CTmax in open vs closed habitats
- Narrow variance in warmer places could mean more stabilizing selection
-
Try variance partitioning CTmax into Hsp, local environment, and phylogenetics
Make CTmax vs Tmax figures with overlay of habitat type.
- regress against latitude and PCA of climate variables too
try framing in terms of integrating proximal and ultimate explanations
put rxn norms in better context of theory; what is the alternative to hotter is better?
- Frazier et al. 2006, AmNat; the alternative is shifts in rxn norm horizontally, but not vertically= perfect-compensation hypothesis. In other words, biochemical adaptation can overcome rate-limiting effects of low temperature so that rmax is independent of Topt. Not mentioned in this explanation is that there can be constraints at higher temperatures that can potentially cause this pattern.
1. among colony variance
ddply(Aph.dat,.(habitat_v2),summarize,CTmax=mean(KO_temp_worker),var=var(KO_temp_worker)) habitat_v2 CTmax var1 deciduous forest 41.04248 0.94437242 flat woods 42.77917 0.1750000PCA of cliamte variables
bclim<-princomp(scale(cbind(Aph.dat[,21:39])))summary(bclim)Importance of components: Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6 Comp.7Standard deviation 3.6328923 1.7748683 1.19556867 0.77430677 0.46454501 0.335626591 0.215453516Proportion of Variance 0.7016431 0.1674725 0.07599067 0.03187406 0.01147273 0.005988581 0.002467848Cumulative Proportion 0.7016431 0.8691156 0.94510623 0.97698029 0.98845302 0.994441598 0.996909446knitr::kable(round(bclim$loadings[,1:2],3))| Comp.1 | Comp.2 | |
|---|---|---|
| bio1 | -0.269 | -0.035 |
| bio2 | -0.144 | -0.354 |
| bio3 | -0.268 | -0.059 |
| bio4 | 0.271 | 0.015 |
| bio5 | -0.249 | -0.102 |
| bio6 | -0.267 | -0.029 |
| bio7 | 0.267 | -0.013 |
| bio8 | -0.214 | -0.040 |
| bio9 | -0.265 | -0.073 |
| bio10 | -0.258 | -0.061 |
| bio11 | -0.270 | -0.034 |
| bio12 | -0.231 | -0.123 |
| bio13 | -0.230 | 0.171 |
| bio14 | 0.078 | -0.495 |
| bio15 | -0.215 | 0.319 |
| bio16 | -0.238 | 0.148 |
| bio17 | 0.058 | -0.514 |
| bio18 | -0.248 | 0.145 |
| bio19 | -0.145 | -0.385 |
regression models; taking first two pcas that explain 86% of variation
pcmod<-lm(KO_temp_worker~Comp.1*habitat_v2+Comp.2*habitat_v2 ,data=Aph.dat)summary(stepAIC(pcmod,direction="both"))Call:lm(formula = KO_temp_worker ~ Comp.1 + habitat_v2 + Comp.2, data = Aph.dat)Residuals: Min 1Q Median 3Q Max -4.0136 -0.3372 0.1448 0.5228 1.5893 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 41.06999 0.10129 405.476 < 2e-16 ***Comp.1 -0.04962 0.03006 -1.651 0.1020 habitat_v2flat woods 1.56474 0.30657 5.104 1.68e-06 ***Comp.2 -0.09366 0.05213 -1.797 0.0755 . ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8862 on 96 degrees of freedomMultiple R-squared: 0.3797, Adjusted R-squared: 0.3603 F-statistic: 19.59 on 3 and 96 DF, p-value: 5.466e-10regressions with Tmax, habitat
xxxxxxxxxxumod<-lm(KO_temp_worker~bio5*habitat_v2 ,data=Aph.dat)summary(stepAIC(umod,direction="both"))Call:lm(formula = KO_temp_worker ~ bio5 + habitat_v2, data = Aph.dat)Residuals: Min 1Q Median 3Q Max -3.8297 -0.3348 0.2332 0.5586 1.4826 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 37.237343 1.084085 34.349 < 2e-16 ***bio5 0.012855 0.003649 3.523 0.000652 ***habitat_v2flat woods 1.376747 0.255980 5.378 5.2e-07 ***---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8605 on 97 degrees of freedomMultiple R-squared: 0.4091, Adjusted R-squared: 0.3969 F-statistic: 33.58 on 2 and 97 DF, p-value: 8.27e-12Figure

regression with MAT
xxxxxxxxxxumod<-lm(KO_temp_worker~bio1*habitat_v2 ,data=Aph.dat)summary(stepAIC(umod,direction="both"))lm(formula = KO_temp_worker ~ bio1 * habitat_v2, data = Aph.dat)Residuals: Min 1Q Median 3Q Max -3.8808 -0.2948 0.1394 0.5549 1.6231 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 40.289262 0.266504 151.177 < 2e-16 ***bio1 0.006325 0.002090 3.027 0.00317 ** habitat_v2flat woods 4.264228 2.013656 2.118 0.03679 * bio1:habitat_v2flat woods -0.015722 0.010713 -1.468 0.14549 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8744 on 96 degrees of freedomMultiple R-squared: 0.3962, Adjusted R-squared: 0.3773 F-statistic: 20.99 on 3 and 96 DF, p-value: 1.534e-10regression with lattitude
xxxxxxxxxxlatmod<-lm(KO_temp_worker~lat*habitat_v2 ,data=Aph.dat)summary(stepAIC(latmod,direction="both"))Call:lm(formula = KO_temp_worker ~ lat * habitat_v2, data = Aph.dat)Residuals: Min 1Q Median 3Q Max -3.9251 -0.2851 0.1050 0.5593 1.6421 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 43.26209 0.80838 53.517 < 2e-16 ***lat -0.05748 0.02079 -2.765 0.00682 ** habitat_v2flat woods -2.95972 2.90928 -1.017 0.31155 lat:habitat_v2flat woods 0.13632 0.09109 1.497 0.13777 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8807 on 96 degrees of freedomMultiple R-squared: 0.3874, Adjusted R-squared: 0.3682 F-statistic: 20.23 on 3 and 96 DF, p-value: 3.043e-10Hsps; pcas and variance partitioning of CTmax
xxxxxxxxxxsummary(pchsp)Importance of components: Comp.1 Comp.2 Comp.3 Comp.4 Comp.5 Comp.6Standard deviation 2.1385967 1.3517804 1.07592411 1.00232658 0.84659220 0.84649220Proportion of Variance 0.3906613 0.1560828 0.09887942 0.08581459 0.06121969 0.06120523Cumulative Proportion 0.3906613 0.5467441 0.64562350 0.73143809 0.79265778 0.85386301knitr::kable(round(pchsp$loadings[,1:7],3))| Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 | Comp.6 | Comp.7 | |
|---|---|---|---|---|---|---|---|
| hsc70 | -0.073 | -0.596 | 0.071 | -0.224 | -0.217 | 0.055 | 0.131 |
| hsp83 | -0.023 | -0.593 | -0.008 | 0.098 | 0.293 | 0.292 | 0.428 |
| hsp40 | -0.023 | 0.008 | 0.461 | 0.803 | -0.159 | 0.237 | -0.098 |
| FC_hsc701468max | -0.321 | -0.160 | 0.404 | -0.273 | -0.043 | -0.006 | -0.451 |
| FC_hsc701468slope | -0.280 | -0.286 | 0.217 | 0.189 | 0.130 | -0.629 | -0.008 |
| FC_hsc701468Tm | -0.374 | 0.157 | 0.226 | -0.133 | -0.245 | -0.283 | 0.247 |
| FC_hsp40541max | -0.350 | -0.082 | -0.324 | 0.129 | -0.097 | 0.273 | -0.358 |
| FC_hsp40541slope | -0.292 | -0.149 | -0.524 | 0.171 | -0.167 | -0.170 | -0.242 |
| FC_hsp40541Tm | -0.368 | 0.063 | -0.260 | 0.149 | -0.323 | 0.130 | 0.355 |
| FC_Hsp83279max | -0.350 | 0.057 | 0.153 | -0.213 | 0.353 | 0.440 | -0.207 |
| FC_Hsp83279slope | -0.290 | 0.171 | -0.145 | 0.186 | 0.694 | -0.167 | 0.129 |
| FC_Hsp83279Tm | -0.351 | 0.310 | 0.171 | -0.143 | -0.119 | 0.194 | 0.393 |
Some stats
xxxxxxxxxxsummary(lm(jj$KO_temp_worker~pchsp$scores[,1]+pchsp$scores[,2]+pchsp$scores[,3]))Call:lm(formula = jj$KO_temp_worker ~ pchsp$scores[, 1] + pchsp$scores[, 2] + pchsp$scores[, 3])Residuals: Min 1Q Median 3Q Max -1.15358 -0.37044 0.04846 0.34646 1.54100 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 41.570122 0.098692 421.211 < 2e-16 ***pchsp$scores[, 1] -0.242155 0.046148 -5.247 6.55e-06 ***pchsp$scores[, 2] -0.001745 0.073009 -0.024 0.981 pchsp$scores[, 3] 0.121858 0.091727 1.328 0.192 ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.6319 on 37 degrees of freedomMultiple R-squared: 0.4419, Adjusted R-squared: 0.3967 F-statistic: 9.767 on 3 and 37 DF, p-value: 6.991e-05
Variance partitioning
xxxxxxxxxxvar10<- varpart(jj$KO_temp_worker, ~ Axis.1 + Axis.2+ Axis.3+ Axis.4+Axis.5+Axis.6+Axis.7+Axis.8+Axis.9, ~bio1+bio5+habitat_v2,~Hsppc1+Hsppc2,data=nw)var10plot(var10)Partition of variance in RDA Call: varpart(Y = jj$KO_temp_worker, X = ~Axis.1 + Axis.2 + Axis.3 + Axis.4 +Axis.5 + Axis.6 + Axis.7 + Axis.8 + Axis.9, ~bio1 + bio5 + habitat_v2, ~Hsppc1+ Hsppc2, data = nw)Explanatory tables:X1: ~Axis.1 + Axis.2 + Axis.3 + Axis.4 + Axis.5 + Axis.6 + Axis.7 + Axis.8 + Axis.9X2: ~bio1 + bio5 + habitat_v2X3: ~Hsppc1 + Hsppc2 No. of explanatory tables: 3 Total variation (SS): 26.477 Variance: 0.66191 No. of observations: 41 Partition table: Df R.square Adj.R.square Testable[a+d+f+g] = X1 9 0.72027 0.63906 TRUE[b+d+e+g] = X2 3 0.64967 0.62126 TRUE[c+e+f+g] = X3 2 0.41531 0.38454 TRUE[a+b+d+e+f+g] = X1+X2 12 0.78605 0.69435 TRUE[a+c+d+e+f+g] = X1+X3 11 0.76028 0.66936 TRUE[b+c+d+e+f+g] = X2+X3 5 0.67973 0.63398 TRUE[a+b+c+d+e+f+g] = All 14 0.80893 0.70604 TRUEIndividual fractions [a] = X1 | X2+X3 9 0.07206 TRUE[b] = X2 | X1+X3 3 0.03668 TRUE[c] = X3 | X1+X2 2 0.01169 TRUE[d] 0 0.21275 FALSE[e] 0 0.01861 FALSE[f] 0 0.00103 FALSE[g] 0 0.35322 FALSE[h] = Residuals 0.29396 FALSEControlling 1 table X [a+d] = X1 | X3 9 0.28482 TRUE[a+f] = X1 | X2 9 0.07309 TRUE[b+d] = X2 | X3 3 0.24944 TRUE[b+e] = X2 | X1 3 0.05529 TRUE[c+e] = X3 | X1 2 0.03029 TRUE[c+f] = X3 | X2 2 0.01271 TRUE---Use function ‘rda’ to test significance of fractions of interest
Slightly better figure

Page 100: 2016-11-14 & 2016-11-15. climate cascade meeting
Project updates:
Hsp gene expression + Ctmax project:
- figure 3, SHC says to switch back branches
- Wrote up methods and results-- go over with Nick then send to SHC
- Submit to PNAS
Multiple stressors ms:
- submitted 2016-10-24 ; in review 2016-11-01
- submitted 2016-10-24 ; in review 2016-11-01
Range limits ms: SHC lab gave verbal edit, still need to incorporate
Thermal niche ms: Lacey and I working on discussion...eta?
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Support with Suiter Prize! $1,000
Thesis related FORMS FOUND HERE
Formatting:
Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- started outline
- started outline
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 101: 2016-11-16Hsp reaction norm stats; adding quadratic term
xxxxxxxxxxlm(formula = KO_temp_worker ~ bio5 + habitat_v2 + I(bio5^2), data = Aph.dat)Residuals: Min 1Q Median 3Q Max -3.6123 -0.3293 0.1297 0.4772 1.8485 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -4.4102626 12.5885230 -0.350 0.726851 bio5 0.2990131 0.0862737 3.466 0.000792 ***habitat_v2flat woods 1.5151487 0.2472431 6.128 1.96e-08 ***I(bio5^2) -0.0004877 0.0001469 -3.320 0.001275 ** ---Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1Residual standard error: 0.8192 on 96 degrees of freedomMultiple R-squared: 0.47, Adjusted R-squared: 0.4534 F-statistic: 28.37 on 3 and 96 DF, p-value: 3.191e-13Page 102: 2016-11-22. climate cascade to do list
Project updates:
Hsp gene expression + Ctmax project:
- rewrite results, intro and send out to NJG and SHC
- Submit to PNAS
- rewrite results, intro and send out to NJG and SHC
Multiple stressors ms:
- major revisions
Range limits ms: SHC lab gave verbal edit, still need to incorporate
Thermal niche ms: In my hands, get to it mid-december
actionable items:
- recheck stats
- recheck figures
- make transitions between paragraphs in discussion
- recheck stats
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (December 1 2016 in SHC lab meeting ; Decemeber 7 2016 in EEEB)
- Support with Suiter Prize! $1,000
Thesis related FORMS FOUND HERE
Formatting:
Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- started outline
- started outline
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 103: 2016-12-06. climate cascade update
Project updates:
Hsp gene expression + Ctmax project:
- rewrite results, intro and send out to NJG and SHC (methods done)
- Submit to PNAS
- rewrite results, intro and send out to NJG and SHC (methods done)
Multiple stressors ms:
major revisions; addressing now
- go over figures
- go over figures
Range limits ms: SHC lab gave verbal edit, still need to incorporate
Thermal niche ms: In my hands, get to it mid-december
actionable items:
- recheck stats (are we using same dataset?)
- recheck figures
- make transitions between paragraphs in discussion (constructing outline)
- recheck stats (are we using same dataset?)
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Attending SICB - Jan 3-8 New Orleans, Give a talk about range limits paper.
- Practice talks: (Decemeber 7 2016 in EEEB)
- Support with Suiter Prize! $1,000
Thesis related FORMS FOUND HERE
Formatting:
Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- started outline
- started outline
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 104: 2016-12-19. climate cascade update
Project updates:
Hsp gene expression + Ctmax project:
- rewrite results, intro and send out to NJG and SHC (methods done)
- Submit to PNAS
- rewrite results, intro and send out to NJG and SHC (methods done)
Multiple stressors ms:
- sent SHC revisions last week
Range limits ms: SHC lab gave verbal edit, still need to incorporate
- Thermal niche ms: Send new draft to Lacy tomorrow.
Stressed in nature MS: Samples to rerun.
- update: Curtis can no longer work+ write on project
- There are 74 samples: 3 days of RNA isolation + cDNA synthesis. 4 gene targets ran in duplicates is 2 plates per gene = 8 plates total. 2 days for 8 plates.
- update: Curtis can no longer work+ write on project
Proteome stability project:
~130 proteins for rudis, ~250 proteins for pogos(we got 500 proteins last time); labelling is ok
- Rerun mass spec, but loading more proteins (Bethany)
Attending SICB - Jan 3-8 New Orleans, on range limits paper.
- SICB talk Jan 8 2017, Sunday, 11:45AM.
- SICB talk Jan 8 2017, Sunday, 11:45AM.
Thesis related FORMS FOUND HERE
Formatting:
Introduction (> 3 pages), manuscripts, then synthesis/conclusion (~3 pages) ; SHC and NJG agree
- started outline
- started outline
- Intent to graduate: February 1st for May.
- Send defense committe form to grad college---now
- Graduate college format check March 4th
- Defense notice 3 weeks before defense (oral defense by March 24th).
- Final thesis April 7th.
- Intent to graduate: February 1st for May.
Page 105: 2016-12-20. Reading a few papers
Reading some papers:
There is a cool paper by Gilchrist and Huey 2001, Evolution, that looks at the cross-generational effect of temperature on fitness in fruit flies. Ones reared from higher temperatures had offspring with higher fitness. This fitness benefit was gained by speeding up development.
- Gilchrist GW, Huey RB (2001) PARENTAL AND DEVELOPMENTAL TEMPERATURE EFFECTS ON THE THERMAL DEPENDENCE OF FITNESS IN DROSOPHILA MELANOGASTER. Evolution 55:209–214. doi: 10.1111/j.0014-3820.2001.tb01287.x
- Gilchrist GW, Huey RB (2001) PARENTAL AND DEVELOPMENTAL TEMPERATURE EFFECTS ON THE THERMAL DEPENDENCE OF FITNESS IN DROSOPHILA MELANOGASTER. Evolution 55:209–214. doi: 10.1111/j.0014-3820.2001.tb01287.x
Cool paper by Huey and Slatkin 1976, The Quarterly Review of Biology which developed the first thermoregulation model in lizards. They construct a mathematical model to quantify the costs and benefits of thermoregulation.
Huey RB, Slatkin M (1976) Cost and Benefits of Lizard Thermoregulation. The Quarterly Review of Biology 51:363–384.
Other follow up models:
- Vickers M, Manicom C, Schwarzkopf L (2011) Extending the cost-benefit model of
thermoregulation: High-temperature environments. Am Nat 177(4):452–461.
- Christian KA, Tracy CR, Tracy CR (2006) Evaluating thermoregulation in reptiles: An
appropriate null model. Am Nat 168(3):421–430.
- Sears MW, Angilletta MJ, Schuler MS, et al (2016) Configuration of the thermal landscape determines thermoregulatory performance of ectotherms. PNAS 201604824. doi: 10.1073/pnas.1604824113 (previous citations 1 and 2 found from this citation) link
- Vickers M, Manicom C, Schwarzkopf L (2011) Extending the cost-benefit model of
thermoregulation: High-temperature environments. Am Nat 177(4):452–461.
- One of Huey's Science papers that shows different populations from 3 continents track chromosomal changes with climate change.
Balanyá J, Oller JM, Huey RB, et al (2006) Global Genetic Change Tracks Global Climate Warming in Drosophila subobscura. Science 313:1773–1775. doi: 10.1126/science.1131002